Virulence effector SidJ evolution in Legionella pneumophila is driven by positive selection and intragenic recombination
- PMID: 34458026
- PMCID: PMC8378335
- DOI: 10.7717/peerj.12000
Virulence effector SidJ evolution in Legionella pneumophila is driven by positive selection and intragenic recombination
Abstract
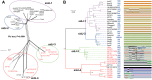
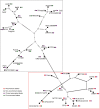
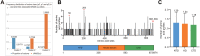
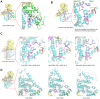
Effector proteins translocated by the Dot/Icm type IV secretion system determine the virulence of Legionella pneumophila (L. pneumophila). Among these effectors, members of the SidE family (SidEs) regulate several cellular processes through a unique phosphoribosyl ubiquitination mechanism mediated by another effector, SidJ. Host-cell calmodulin (CaM) activates SidJ to glutamylate the SidEs of ubiquitin (Ub) ligases and to make a balanced Ub ligase activity. Given the central role of SidJ in this regulatory process, studying the nature of evolution of sidJ is important to understand the virulence of L. pneumophila and the interaction between the bacteria and its hosts. By studying sidJ from a large number of L. pneumophila strains and using various molecular evolution algorithms, we demonstrated that intragenic recombination drove the evolution of sidJ and contributed to sidJ diversification. Additionally, we showed that four codons of sidJ which are located in the N-terminal (NTD) (codons 58 and 200) and C-terminal (CTD) (codons 868 and 869) domains, but not in the kinase domain (KD) had been subjected to strong positive selection pressure, and variable mutation profiles of these codons were identified. Protein structural modeling of SidJ provided possible explanations for these mutations. Codons 868 and 869 mutations might engage in regulating the interactions of SidJ with CaM through hydrogen bonds and affect the CaM docking to SidJ. Mutation in codon 58 of SidJ might affect the distribution of main-chain atoms that are associated with the interaction with CaM. In contrast, mutations in codon 200 might influence the α-helix stability in the NTD. These mutations might be important to balance Ub ligase activity for different L. pneumophila hosts. This study first reported that intragenic recombination and positive Darwinian selection both shaped the genetic plasticity of sidJ, contributing to a deeper understanding of the adaptive mechanisms of this intracellular bacterium to different hosts.
Keywords: Adaptive evolution; Evolution; Intragenic recombination; Legionella pneumophila; Positive selection; SidJ; Virulence effector.
©2021 Zhan et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



References
-
- Al-Khodor S, Price CT, Habyarimana F, Kalia A, Abu Kwaik Y. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Molecular Microbiology. 2008;70:908–923. doi: 10.1111/j.1365-2958.2008.06453.x. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

