Class I PI3K Provide Lipid Substrate in T Cell Autophagy Through Linked Activity of Inositol Phosphatases
- PMID: 34458267
- PMCID: PMC8397451
- DOI: 10.3389/fcell.2021.709398
Class I PI3K Provide Lipid Substrate in T Cell Autophagy Through Linked Activity of Inositol Phosphatases
Abstract
Autophagy, a highly conserved intracellular process, has been identified as a novel mechanism regulating T lymphocyte homeostasis. Herein, we demonstrate that both starvation- and T cell receptor-mediated autophagy induction requires class I phosphatidylinositol-3 kinases to produce PI(3)P. In contrast, common gamma chain cytokines are suppressors of autophagy despite their ability to activate the PI3K pathway. T cells lacking the PI3KI regulatory subunits, p85 and p55, were almost completely unable to activate TCR-mediated autophagy and had concurrent defects in PI(3)P production. Additionally, T lymphocytes upregulate polyinositol phosphatases in response to autophagic stimuli, and the activity of the inositol phosphatases Inpp4 and SHIP are required for TCR-mediated autophagy induction. Addition of exogenous PI(3,4)P2 can supplement cellular PI(3)P and accelerate the outcome of activation-induced autophagy. TCR-mediated autophagy also requires internalization of the TCR complex, suggesting that this kinase/phosphatase activity is localized in internalized vesicles. Finally, HIV-induced bystander CD4+ T cell autophagy is dependent upon PI3KI. Overall, our data elucidate an important pathway linking TCR activation to autophagy, via induction of PI3KI activity and inositol phosphatase upregulation to produce PI(3)P.
Keywords: HIV; PI3K I; T cells; autophagy; cytokines; lipid kinase; lipid phosphatase.
Copyright © 2021 McLeod, Saxena, Carico and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
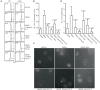

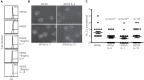
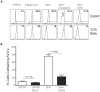
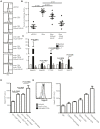

References
-
- Axe E. L., Walker S. A., Manifava M., Chandra P., Roderick H. L., Habermann A., et al. (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182 685–701. 10.1083/jcb.200803137 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

