Metathramycin, a new bioactive aureolic acid discovered by heterologous expression of a metagenome derived biosynthetic pathway
- PMID: 34458799
- PMCID: PMC8341913
- DOI: 10.1039/d0cb00228c
Metathramycin, a new bioactive aureolic acid discovered by heterologous expression of a metagenome derived biosynthetic pathway
Abstract
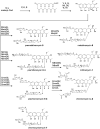
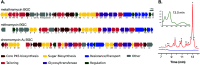
Bacterial natural products have been a rich source of bioactive compounds for drug development, and advances in DNA sequencing, informatics and molecular biology have opened new avenues for their discovery. Here, we describe the isolation of an aureolic acid biosynthetic gene cluster from a metagenome library derived from a New Zealand soil sample. Heterologous expression of this pathway in Streptomyces albus resulted in the production and isolation of two new aureolic acid compounds, one of which (metathramycin, 6) possesses potent bioactivity against a human colon carcinoma cell line (HCT-116, IC50 = 14.6 nM). As metathramycin was a minor constituent of the fermentation extract, its discovery relied on a combination of approaches including bioactivity guided fractionation, MS/MS characterisation and pathway engineering. This study not only demonstrates the presence of previously uncharacterised aureolic acids in the environment, but also the value of an integrated natural product discovery approach which may be generally applicable to low abundance bioactive metabolites.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



References
-
- Menéndez N. Nur-e-Alam M. Braña A. F. Rohr J. Salas J. A. Méndez C. Chem. Biol. 2004;11:21–32. - PubMed
-
- Sabín J. G. Biochem. Pharmacol. Open Access. 2013;2:1–3.
LinkOut - more resources
Full Text Sources
Research Materials

