Conservation and divergence of meiotic DNA double strand break forming mechanisms in Arabidopsis thaliana
- PMID: 34458909
- PMCID: PMC8464057
- DOI: 10.1093/nar/gkab715
Conservation and divergence of meiotic DNA double strand break forming mechanisms in Arabidopsis thaliana
Abstract
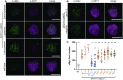
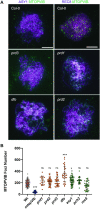
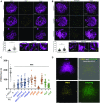
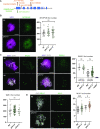
In the current meiotic recombination initiation model, the SPO11 catalytic subunits associate with MTOPVIB to form a Topoisomerase VI-like complex that generates DNA double strand breaks (DSBs). Four additional proteins, PRD1/AtMEI1, PRD2/AtMEI4, PRD3/AtMER2 and the plant specific DFO are required for meiotic DSB formation. Here we show that (i) MTOPVIB and PRD1 provide the link between the catalytic sub-complex and the other DSB proteins, (ii) PRD3/AtMER2, while localized to the axis, does not assemble a canonical pre-DSB complex but establishes a direct link between the DSB-forming and resection machineries, (iii) DFO controls MTOPVIB foci formation and is part of a divergent RMM-like complex including PHS1/AtREC114 and PRD2/AtMEI4 but not PRD3/AtMER2, (iv) PHS1/AtREC114 is absolutely unnecessary for DSB formation despite having a conserved position within the DSB protein network and (v) MTOPVIB and PRD2/AtMEI4 interact directly with chromosome axis proteins to anchor the meiotic DSB machinery to the axis.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M.. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 2015; 66:297–327. - PubMed
-
- Zelkowski M., Olson M.A., Wang M., Pawlowski W.P.. Diversity and determinants of meiotic recombination landscapes. Trends Genet. 2019; 35:359–370. - PubMed
-
- de Massy B.Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 2013; 47:563–599. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

