Inhibition of the cGAS-STING pathway ameliorates the premature senescence hallmarks of Ataxia-Telangiectasia brain organoids
- PMID: 34459078
- PMCID: PMC8441292
- DOI: 10.1111/acel.13468
Inhibition of the cGAS-STING pathway ameliorates the premature senescence hallmarks of Ataxia-Telangiectasia brain organoids
Abstract
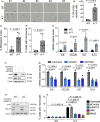
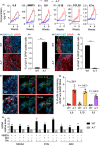
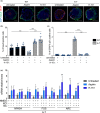
Ataxia-telangiectasia (A-T) is a genetic disorder caused by the lack of functional ATM kinase. A-T is characterized by chronic inflammation, neurodegeneration and premature ageing features that are associated with increased genome instability, nuclear shape alterations, micronuclei accumulation, neuronal defects and premature entry into cellular senescence. The causal relationship between the detrimental inflammatory signature and the neurological deficiencies of A-T remains elusive. Here, we utilize human pluripotent stem cell-derived cortical brain organoids to study A-T neuropathology. Mechanistically, we show that the cGAS-STING pathway is required for the recognition of micronuclei and induction of a senescence-associated secretory phenotype (SASP) in A-T olfactory neurosphere-derived cells and brain organoids. We further demonstrate that cGAS and STING inhibition effectively suppresses self-DNA-triggered SASP expression in A-T brain organoids, inhibits astrocyte senescence and neurodegeneration, and ameliorates A-T brain organoid neuropathology. Our study thus reveals that increased cGAS and STING activity is an important contributor to chronic inflammation and premature senescence in the central nervous system of A-T and constitutes a novel therapeutic target for treating neuropathology in A-T patients.
Keywords: Ataxia-Telangiectasia; brain aging; brain organoids; cGAS-STING signalling; cellular senescence; neurodegeneration.
© 2021 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Acosta, J. C., Banito, A., Wuestefeld, T., Georgilis, A., Janich, P., Morton, J. P., Athineos, D., Kang, T.‐W., Lasitschka, F., Andrulis, M., Pascual, G., Morris, K. J., Khan, S., Jin, H., Dharmalingam, G., Snijders, A. P., Carroll, T., Capper, D., Pritchard, C., … Gil, J. (2013). A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nature Cell Biology, 15(8), 978–990. 10.1038/ncb2784 - DOI - PMC - PubMed
-
- Aguado, J., Sola‐Carvajal, A., Cancila, V., Revêchon, G., Ong, P. F., Jones‐Weinert, C. W., Wallén Arzt, E., Lattanzi, G., Dreesen, O., Tripodo, C., Rossiello, F., Eriksson, M., & d’Adda di Fagagna, F. (2019). Inhibition of DNA damage response at telomeres improves the detrimental phenotypes of Hutchinson‐Gilford Progeria Syndrome. Nature Communications, 10(1), 4990. 10.1038/s41467-019-13018-3 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

