Helios represses megakaryocyte priming in hematopoietic stem and progenitor cells
- PMID: 34459852
- PMCID: PMC8406645
- DOI: 10.1084/jem.20202317
Helios represses megakaryocyte priming in hematopoietic stem and progenitor cells
Abstract
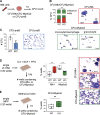
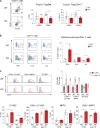
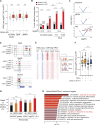
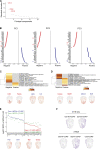
Our understanding of cell fate decisions in hematopoietic stem cells is incomplete. Here, we show that the transcription factor Helios is highly expressed in murine hematopoietic stem and progenitor cells (HSPCs), where it is required to suppress the separation of the platelet/megakaryocyte lineage from the HSPC pool. Helios acts mainly in quiescent cells, where it directly represses the megakaryocyte gene expression program in cells as early as the stem cell stage. Helios binding promotes chromatin compaction, notably at the regulatory regions of platelet-specific genes recognized by the Gata2 and Runx1 transcriptional activators, implicated in megakaryocyte priming. Helios null HSPCs are biased toward the megakaryocyte lineage at the expense of the lymphoid and partially resemble cells of aging animals. We propose that Helios acts as a guardian of HSPC pluripotency by continuously repressing the megakaryocyte fate, which in turn allows downstream lymphoid priming to take place. These results highlight the importance of negative and positive priming events in lineage commitment.
© 2021 Cova et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

