Fully 3D Active Surface with Machine Learning for PET Image Segmentation
- PMID: 34460557
- PMCID: PMC8321170
- DOI: 10.3390/jimaging6110113
Fully 3D Active Surface with Machine Learning for PET Image Segmentation
Abstract
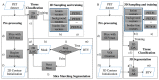
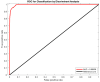
In order to tackle three-dimensional tumor volume reconstruction from Positron Emission Tomography (PET) images, most of the existing algorithms rely on the segmentation of independent PET slices. To exploit cross-slice information, typically overlooked in these 2D implementations, I present an algorithm capable of achieving the volume reconstruction directly in 3D, by leveraging an active surface algorithm. The evolution of such surface performs the segmentation of the whole stack of slices simultaneously and can handle changes in topology. Furthermore, no artificial stop condition is required, as the active surface will naturally converge to a stable topology. In addition, I include a machine learning component to enhance the accuracy of the segmentation process. The latter consists of a forcing term based on classification results from a discriminant analysis algorithm, which is included directly in the mathematical formulation of the energy function driving surface evolution. It is worth noting that the training of such a component requires minimal data compared to more involved deep learning methods. Only eight patients (i.e., two lung, four head and neck, and two brain cancers) were used for training and testing the machine learning component, while fifty patients (i.e., 10 lung, 25 head and neck, and 15 brain cancers) were used to test the full 3D reconstruction algorithm. Performance evaluation is based on the same dataset of patients discussed in my previous work, where the segmentation was performed using the 2D active contour. The results confirm that the active surface algorithm is superior to the active contour algorithm, outperforming the earlier approach on all the investigated anatomical districts with a dice similarity coefficient of 90.47 ± 2.36% for lung cancer, 88.30 ± 2.89% for head and neck cancer, and 90.29 ± 2.52% for brain cancer. Based on the reported results, it can be claimed that the migration into a 3D system yielded a practical benefit justifying the effort to rewrite an existing 2D system for PET imaging segmentation.
Keywords: 3D segmentation; PET imaging; active surface; discriminant analysis; machine learning.
Conflict of interest statement
The author declares no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

