Accelerating 3D Medical Image Segmentation by Adaptive Small-Scale Target Localization
- PMID: 34460634
- PMCID: PMC8321270
- DOI: 10.3390/jimaging7020035
Accelerating 3D Medical Image Segmentation by Adaptive Small-Scale Target Localization
Abstract
The prevailing approach for three-dimensional (3D) medical image segmentation is to use convolutional networks. Recently, deep learning methods have achieved human-level performance in several important applied problems, such as volumetry for lung-cancer diagnosis or delineation for radiation therapy planning. However, state-of-the-art architectures, such as U-Net and DeepMedic, are computationally heavy and require workstations accelerated with graphics processing units for fast inference. However, scarce research has been conducted concerning enabling fast central processing unit computations for such networks. Our paper fills this gap. We propose a new segmentation method with a human-like technique to segment a 3D study. First, we analyze the image at a small scale to identify areas of interest and then process only relevant feature-map patches. Our method not only reduces the inference time from 10 min to 15 s but also preserves state-of-the-art segmentation quality, as we illustrate in the set of experiments with two large datasets.
Keywords: computed tomography (CT); deep learning; magnetic resonance imaging (MRI); medical image segmentation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
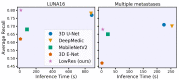
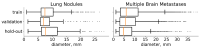

References
-
- Greenspan H., Van Ginneken B., Summers R.M. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging. 2016;35:1153–1159. doi: 10.1109/TMI.2016.2553401. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

