A multi-objective genetic algorithm to find active modules in multiplex biological networks
- PMID: 34460810
- PMCID: PMC8452006
- DOI: 10.1371/journal.pcbi.1009263
A multi-objective genetic algorithm to find active modules in multiplex biological networks
Abstract
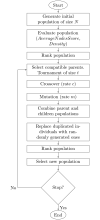
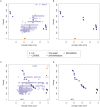
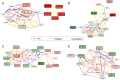
The identification of subnetworks of interest-or active modules-by integrating biological networks with molecular profiles is a key resource to inform on the processes perturbed in different cellular conditions. We here propose MOGAMUN, a Multi-Objective Genetic Algorithm to identify active modules in MUltiplex biological Networks. MOGAMUN optimizes both the density of interactions and the scores of the nodes (e.g., their differential expression). We compare MOGAMUN with state-of-the-art methods, representative of different algorithms dedicated to the identification of active modules in single networks. MOGAMUN identifies dense and high-scoring modules that are also easier to interpret. In addition, to our knowledge, MOGAMUN is the first method able to use multiplex networks. Multiplex networks are composed of different layers of physical and functional relationships between genes and proteins. Each layer is associated to its own meaning, topology, and biases; the multiplex framework allows exploiting this diversity of biological networks. We applied MOGAMUN to identify cellular processes perturbed in Facio-Scapulo-Humeral muscular Dystrophy, by integrating RNA-seq expression data with a multiplex biological network. We identified different active modules of interest, thereby providing new angles for investigating the pathomechanisms of this disease. Availability: MOGAMUN is available at https://github.com/elvanov/MOGAMUN and as a Bioconductor package at https://bioconductor.org/packages/release/bioc/html/MOGAMUN.html. Contact: anais.baudot@univ-amu.fr.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

