IFN-γ signature in the plasma proteome distinguishes pediatric hemophagocytic lymphohistiocytosis from sepsis and SIRS
- PMID: 34461635
- PMCID: PMC8525230
- DOI: 10.1182/bloodadvances.2021004287
IFN-γ signature in the plasma proteome distinguishes pediatric hemophagocytic lymphohistiocytosis from sepsis and SIRS
Abstract
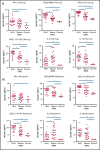
Hemophagocytic lymphohistiocytosis (HLH) is a syndrome characterized by pathologic immune activation in which prompt recognition and initiation of immune suppression is essential for survival. Children with HLH have many overlapping clinical features with critically ill children with sepsis and systemic inflammatory response syndrome (SIRS) in whom alternative therapies are indicated. To determine whether plasma biomarkers could differentiate HLH from other inflammatory conditions and to better define a core inflammatory signature of HLH, concentrations of inflammatory plasma proteins were compared in 40 patients with HLH to 47 pediatric patients with severe sepsis or SIRS. Fifteen of 135 analytes were significantly different in HLH plasma compared with SIRS/sepsis, including increased interferon-γ (IFN-γ)-regulated chemokines CXCL9, CXCL10, and CXCL11. Furthermore, a 2-analyte plasma protein classifier including CXCL9 and interleukin-6 was able to differentiate HLH from SIRS/sepsis. Gene expression in CD8+ T cells and activated monocytes from blood were also enriched for IFN-γ pathway signatures in peripheral blood cells from patients with HLH compared with SIRS/sepsis. This study identifies differential expression of inflammatory proteins as a diagnostic strategy to identify critically ill children with HLH, and comprehensive unbiased analysis of inflammatory plasma proteins and global gene expression demonstrates that IFN-γ signaling is uniquely elevated in HLH. In addition to demonstrating the ability of diagnostic criteria for HLH and sepsis or SIRS to identify groups with distinct inflammatory patterns, results from this study support the potential for prospective evaluation of inflammatory biomarkers to aid in diagnosis of and optimizing therapeutic strategies for children with distinctive hyperinflammatory syndromes.
© 2021 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: M.B.J., M.L.H., and C.E.A. have served as consultants for Sobi. M.L.H. serves on a Data Safety Monitoring Committee for Novimmune. All remain authors declare no competing financial interests.
Figures




References
-
- Henter JI, Horne A, Aricó M, et al. . HLH-2004: diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer. 2007;48(2):124-131. - PubMed
-
- Goldstein B, Giroir B, Randolph A; International Consensus Conference on Pediatric Sepsis .International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6(1):2-8. - PubMed
-
- Levy MM, Fink MP, Marshall JC, et al. ; SCCM/ESICM/ACCP/ATS/SIS . 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31(4):1250-1256. - PubMed

