Addiction-Associated Genetic Variants Implicate Brain Cell Type- and Region-Specific Cis-Regulatory Elements in Addiction Neurobiology
- PMID: 34462306
- PMCID: PMC8549541
- DOI: 10.1523/JNEUROSCI.2534-20.2021
Addiction-Associated Genetic Variants Implicate Brain Cell Type- and Region-Specific Cis-Regulatory Elements in Addiction Neurobiology
Abstract
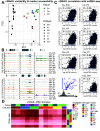
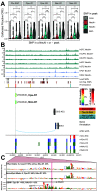
Recent large genome-wide association studies have identified multiple confident risk loci linked to addiction-associated behavioral traits. Most genetic variants linked to addiction-associated traits lie in noncoding regions of the genome, likely disrupting cis-regulatory element (CRE) function. CREs tend to be highly cell type-specific and may contribute to the functional development of the neural circuits underlying addiction. Yet, a systematic approach for predicting the impact of risk variants on the CREs of specific cell populations is lacking. To dissect the cell types and brain regions underlying addiction-associated traits, we applied stratified linkage disequilibrium score regression to compare genome-wide association studies to genomic regions collected from human and mouse assays for open chromatin, which is associated with CRE activity. We found enrichment of addiction-associated variants in putative CREs marked by open chromatin in neuronal (NeuN+) nuclei collected from multiple prefrontal cortical areas and striatal regions known to play major roles in reward and addiction. To further dissect the cell type-specific basis of addiction-associated traits, we also identified enrichments in human orthologs of open chromatin regions of female and male mouse neuronal subtypes: cortical excitatory, D1, D2, and PV. Last, we developed machine learning models to predict mouse cell type-specific open chromatin, enabling us to further categorize human NeuN+ open chromatin regions into cortical excitatory or striatal D1 and D2 neurons and predict the functional impact of addiction-associated genetic variants. Our results suggest that different neuronal subtypes within the reward system play distinct roles in the variety of traits that contribute to addiction.SIGNIFICANCE STATEMENT We combine statistical genetic and machine learning techniques to find that the predisposition to for nicotine, alcohol, and cannabis use behaviors can be partially explained by genetic variants in conserved regulatory elements within specific brain regions and neuronal subtypes of the reward system. Our computational framework can flexibly integrate open chromatin data across species to screen for putative causal variants in a cell type- and tissue-specific manner for numerous complex traits.
Keywords: addiction; deep learning; epigenetics; genomics; machine learning; neural circuits.
Copyright © 2021 Srinivasan et al.
Figures









References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
