LIVECell-A large-scale dataset for label-free live cell segmentation
- PMID: 34462594
- PMCID: PMC8440198
- DOI: 10.1038/s41592-021-01249-6
LIVECell-A large-scale dataset for label-free live cell segmentation
Abstract
Light microscopy combined with well-established protocols of two-dimensional cell culture facilitates high-throughput quantitative imaging to study biological phenomena. Accurate segmentation of individual cells in images enables exploration of complex biological questions, but can require sophisticated imaging processing pipelines in cases of low contrast and high object density. Deep learning-based methods are considered state-of-the-art for image segmentation but typically require vast amounts of annotated data, for which there is no suitable resource available in the field of label-free cellular imaging. Here, we present LIVECell, a large, high-quality, manually annotated and expert-validated dataset of phase-contrast images, consisting of over 1.6 million cells from a diverse set of cell morphologies and culture densities. To further demonstrate its use, we train convolutional neural network-based models using LIVECell and evaluate model segmentation accuracy with a proposed a suite of benchmarks.
© 2021. The Author(s).
Conflict of interest statement
C.E., T.R.J., N.B., T.D., J.T. and R.S. are currently employed by Sartorius that funded the image annotation and provided the Incucyte Live-Cell Analysis system used to acquire the images in LIVECell. The remaining authors declare no competing interests.
Figures

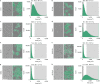


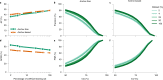
References
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

