Effector differentiation downstream of lineage commitment in ILC1s is driven by Hobit across tissues
- PMID: 34462601
- PMCID: PMC7611762
- DOI: 10.1038/s41590-021-01013-0
Effector differentiation downstream of lineage commitment in ILC1s is driven by Hobit across tissues
Abstract
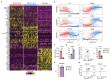
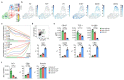
Innate lymphoid cells (ILCs) participate in tissue homeostasis, inflammation, and early immunity against infection. It is unclear how ILCs acquire effector function and whether these mechanisms differ between organs. Through multiplexed single-cell mRNA sequencing, we identified cKit+CD127hiTCF-1hi early differentiation stages of T-bet+ ILC1s. These cells were present across different organs and had the potential to mature toward CD127intTCF-1int and CD127-TCF-1- ILC1s. Paralleling a gradual loss of TCF-1, differentiating ILC1s forfeited their expansion potential while increasing expression of effector molecules, reminiscent of T cell differentiation in secondary lymphoid organs. The transcription factor Hobit was induced in TCF-1hi ILC1s and was required for their effector differentiation. These findings reveal sequential mechanisms of ILC1 lineage commitment and effector differentiation that are conserved across tissues. Our analyses suggest that ILC1s emerge as TCF-1hi cells in the periphery and acquire a spectrum of organ-specific effector phenotypes through a uniform Hobit-dependent differentiation pathway driven by local cues.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
The authors declare no competing interests. K.K. is an employee of Morphosys AG and former employee of the Roche Innovation Center Munich.
Figures














References
-
- Vivier E, et al. Innate Lymphoid Cells: 10 Years On. Cell. 2018;174:1054–1066. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

