This is a preprint.
Global disparities in SARS-CoV-2 genomic surveillance
- PMID: 34462754
- PMCID: PMC8404891
- DOI: 10.1101/2021.08.21.21262393
Global disparities in SARS-CoV-2 genomic surveillance
Update in
-
Global disparities in SARS-CoV-2 genomic surveillance.Nat Commun. 2022 Nov 16;13(1):7003. doi: 10.1038/s41467-022-33713-y. Nat Commun. 2022. PMID: 36385137 Free PMC article.
Abstract
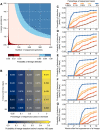
Genomic sequencing provides critical information to track the evolution and spread of SARS-CoV-2, optimize molecular tests, treatments and vaccines, and guide public health responses. To investigate the spatiotemporal heterogeneity in the global SARS-CoV-2 genomic surveillance, we estimated the impact of sequencing intensity and turnaround times (TAT) on variant detection in 167 countries. Most countries submit genomes >21 days after sample collection, and 77% of low and middle income countries sequenced <0.5% of their cases. We found that sequencing at least 0.5% of the cases, with a TAT <21 days, could be a benchmark for SARS-CoV-2 genomic surveillance efforts. Socioeconomic inequalities substantially impact our ability to quickly detect SARS-CoV-2 variants, and undermine the global pandemic preparedness.
Figures

References
-
- WHO. WHO COVID-19 Explorer (2021), (available at https://worldhealthorg.shinyapps.io/covid/).
-
- Lauring A. S., Hodcroft E. B., Genetic Variants of SARS-CoV-2—What Do They Mean? JAMA. 325, 529–531 (2021). - PubMed
-
- CDC, Cases, Data, and Surveillance (2021), (available at https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveill...).
-
- WHO, Tracking SARS-CoV-2 variants. WHO; (2021), (available at https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
