This is a preprint.
TREATMENT WITH SOLUBLE CD24 ATTENUATES COVID-19-ASSOCIATED SYSTEMIC IMMUNOPATHOLOGY
- PMID: 34462760
- PMCID: PMC8404902
- DOI: 10.1101/2021.08.18.21262258
TREATMENT WITH SOLUBLE CD24 ATTENUATES COVID-19-ASSOCIATED SYSTEMIC IMMUNOPATHOLOGY
Update in
-
Treatment with soluble CD24 attenuates COVID-19-associated systemic immunopathology.J Hematol Oncol. 2022 Jan 10;15(1):5. doi: 10.1186/s13045-021-01222-y. J Hematol Oncol. 2022. PMID: 35012610 Free PMC article. Clinical Trial.
Abstract
Background: SARS-CoV-2 causes COVID-19 through direct lysis of infected lung epithelial cells, which releases damage-associated molecular patterns (DAMPs) and induces a pro-inflammatory cytokine milieu causing systemic inflammation. Anti-viral and anti-inflammatory agents have shown limited therapeutic efficacy. Soluble CD24 (CD24Fc) is able to blunt the broad inflammatory response induced by DAMPs in multiple models. A recent randomized phase III trial evaluating the impact of CD24Fc in patients with severe COVID-19 demonstrated encouraging clinical efficacy.
Methods: We studied peripheral blood samples obtained from patients enrolled at a single institution in the SAC-COVID trial (NCT04317040) collected before and after treatment with CD24Fc or placebo. We performed high dimensional spectral flow cytometry analysis of peripheral blood mononuclear cells and measured the levels of a broad array of cytokines and chemokines. A systems analytical approach was used to discern the impact of CD24Fc treatment on immune homeostasis in patients with COVID-19.
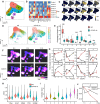
Findings: Twenty-two patients were enrolled, and the clinical characteristics from the CD24Fc vs. placebo groups were matched. Using high-content spectral flow cytometry and network-level analysis, we found systemic hyper-activation of multiple cellular compartments in the placebo group, including CD8+ T cells, CD4+ T cells, and CD56+ NK cells. By contrast, CD24Fc-treated patients demonstrated blunted systemic inflammation, with a return to homeostasis in both NK and T cells within days without compromising the ability of patients to mount an effective anti-Spike protein antibody response. A single dose of CD24Fc significantly attenuated induction of the systemic cytokine response, including expression of IL-10 and IL-15, and diminished the coexpression and network connectivity among extensive circulating inflammatory cytokines, the parameters associated with COVID-19 disease severity.
Interpretation: Our data demonstrates that CD24Fc treatment rapidly down-modulates systemic inflammation and restores immune homeostasis in SARS-CoV-2-infected individuals, supporting further development of CD24Fc as a novel therapeutic against severe COVID-19.
Funding: NIH.
Figures



References
-
- Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021; 596(7871): 276–80. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
