Modeling SARS-CoV-2 proteins in the CASP-commons experiment
- PMID: 34462960
- PMCID: PMC8616790
- DOI: 10.1002/prot.26231
Modeling SARS-CoV-2 proteins in the CASP-commons experiment
Abstract
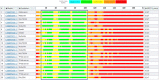
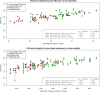
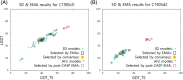
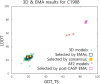
Critical Assessment of Structure Prediction (CASP) is an organization aimed at advancing the state of the art in computing protein structure from sequence. In the spring of 2020, CASP launched a community project to compute the structures of the most structurally challenging proteins coded for in the SARS-CoV-2 genome. Forty-seven research groups submitted over 3000 three-dimensional models and 700 sets of accuracy estimates on 10 proteins. The resulting models were released to the public. CASP community members also worked together to provide estimates of local and global accuracy and identify structure-based domain boundaries for some proteins. Subsequently, two of these structures (ORF3a and ORF8) have been solved experimentally, allowing assessment of both model quality and the accuracy estimates. Models from the AlphaFold2 group were found to have good agreement with the experimental structures, with main chain GDT_TS accuracy scores ranging from 63 (a correct topology) to 87 (competitive with experiment).
Keywords: CASP; COVID; EMA; SARS-CoV-2; model accuracy; protein structure prediction.
© 2021 Wiley Periodicals LLC.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- DE-SC0021303/U.S. Department of Energy
- GM100482/GM/NIGMS NIH HHS/United States
- R35 GM126948/GM/NIGMS NIH HHS/United States
- DBI 1759934/National Science Foundation
- UMO-2017/25/B/ST4/01026/Narodowe Centrum Nauki
- R01 GM123055/GM/NIGMS NIH HHS/United States
- R01GM123055/NH/NIH HHS/United States
- R01 GM100482/GM/NIGMS NIH HHS/United States
- 2020M3A9G7103933/National Research Foundation of Korea
- BB/T018496/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- S-MIP-21-35/Research Council of Lithuania
- T32 GM132024/GM/NIGMS NIH HHS/United States
- CMMI1825941/National Science Foundation
- GA76-11/ICM, University of Warsaw
- unres19/Cyfronet, AGH University of Science and Technology, Cracow
- R01GM133840/NH/NIH HHS/United States
- GM093123/NH/NIH HHS/United States
- UMO-2017/26/M/ST4/00044/Narodowe Centrum Nauki
- BBS/E/W/0012843D/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R35 GM138146/GM/NIGMS NIH HHS/United States
- DE-SC0020400/U.S. Department of Energy
- DBI2003635/National Science Foundation
- IIS1763246/National Science Foundation
- MCB1925643/National Science Foundation
- 2019M3E5D4066898/National Research Foundation of Korea
- R01 GM133840/GM/NIGMS NIH HHS/United States
- R01 GM093123/GM/NIGMS NIH HHS/United States
- S-MIP-17-60/Research Council of Lithuania
- JP20am0101110/Japan Agency for Medical Research and Development
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

