Myxobacterial depsipeptide chondramides interrupt SARS-CoV-2 entry by targeting its broad, cell tropic spike protein
- PMID: 34463219
- PMCID: PMC8436362
- DOI: 10.1080/07391102.2021.1969281
Myxobacterial depsipeptide chondramides interrupt SARS-CoV-2 entry by targeting its broad, cell tropic spike protein
Abstract
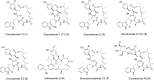
The severity of the COVID-19 pandemic has necessitated the search for drugs against SARS-CoV-2. In this study, we explored via in silico approaches myxobacterial secondary metabolites against various receptor-binding regions of SARS-CoV-2 spike which are responsible in recognition and attachment to host cell receptors mechanisms, namely ACE2, GRP78, and NRP1. In general, cyclic depsipeptide chondramides conferred high affinities toward the spike RBD, showing strong binding to the known viral hot spots Arg403, Gln493 and Gln498 and better selectivity compared to most host cell receptors studied. Among them, chondramide C3 (1) exhibited a binding energy which remained relatively constant when docked against most of the spike variants. Chondramide C (2) on the other hand exhibited strong affinity against spike variants identified in the United Kingdom (N501Y), South Africa (N501Y, E484K, K417N) and Brazil (N501Y, E484K, K417T). Chondramide C6 (9) showed highest BE towards GRP78 RBD. Molecular dynamics simulations were also performed for chondramides 1 and 2 against SARS-CoV-2 spike RBD of the Wuhan wild-type and the South African variant, respectively, where resulting complexes demonstrated dynamic stability within a 120-ns simulation time. Protein-protein binding experiments using HADDOCK illustrated weaker binding affinity for complexed chondramide ligands in the RBD against the studied host cell receptors. The chondramide derivatives in general possessed favorable pharmacokinetic properties, highlighting their potential as prototypic anti-COVID-19 drugs limiting viral attachment and possibly minimizing viral infection.Communicated by Ramaswamy H. Sarma.
Keywords: Antiviral agents; COVID-19; SARS-CoV-2 spike proteins and variants; molecular docking; protein-protein interactions.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures



References
-
- Allam, L., Ghrifi, F., Mohammed, H., El Hafidi, N., El Jaoudi, R., El Harti, J., Lmimouni, B., Belyamani, L., & Ibrahimi, A. (2020). Targeting the GRP78-dependent SARS-CoV-2 cell entry by peptides and small molecules. Bioinformatics and Biology Insights, 14, 117793222096511–117793222096550. 10.1177/1177932220965505 - DOI - PMC - PubMed
-
- Brogi, S., Ramunno, A., Savi, L., Chemi, G., Alfano, G., Pecorelli, A., Pambianchi, E., Galatello, P., Compagnoni, G., Focher, F., Biamonti, G., Valacchi, G., Butini, S., Gemma, S., Campiani, G., & Brindisi, M. (2017). First dual AK/GSK-3β inhibitors endowed with antioxidant properties as multifunctional, potential neuroprotective agents. European Journal of Medicinal Chemistry, 138, 438–457. 10.1016/j.ejmech.2017.06.017 - DOI - PubMed
-
- Cantuti-Castelvetri, L., Ojha, R., Pedro, L. D., Djannatian, M., Franz, J., Kuivanen, S., van der Meer, F., Kallio, K., Kaya, T., Anastasina, M., Smura, T., Levanov, L., Szirovicza, L., Tobi, A., Kallio-Kokko, H., Österlund, P., Joensuu, M., Meunier, F. A., Butcher, S. J., … Simons, M. (2020). Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science (New York, N.Y.), 370(6518), 856–860. 10.1126/science.abd2985 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
