Dominant Bacterial Phyla from the Human Gut Show Widespread Ability To Transform and Conjugate Bile Acids
- PMID: 34463573
- PMCID: PMC12338150
- DOI: 10.1128/mSystems.00805-21
Dominant Bacterial Phyla from the Human Gut Show Widespread Ability To Transform and Conjugate Bile Acids
Abstract
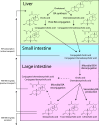
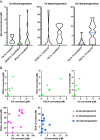
Gut bacteria influence human physiology by chemically modifying host-synthesized primary bile acids. These modified bile acids, known as secondary bile acids, can act as signaling molecules that modulate host lipid, glucose, and energy metabolism and affect gut microbiota composition via selective antimicrobial properties. However, knowledge regarding the bile acid-transforming capabilities of individual gut microbes remains limited. To help address this knowledge gap, we screened 72 bacterial isolates, spanning seven major phyla commonly found in the human gut, for their ability to chemically modify unconjugated bile acids. We found that 43 isolates, representing 41 species, were capable of in vitro modification of one or more of the three most abundant unconjugated bile acids in humans: cholic acid, chenodeoxycholic acid, and deoxycholic acid. Of these, 32 species have not been previously described as bile acid transformers. The most prevalent bile acid transformations detected were oxidation of 3α-, 7α-, or 12α-hydroxyl groups on the steroid core, a reaction catalyzed by hydroxysteroid dehydrogenases. In addition, we found 7α-dehydroxylation activity to be distributed across various bacterial genera, and we observed several other complex bile acid transformations. Finally, our screen revealed widespread bacterial conjugation of primary and secondary bile acids to glycine, a process that was thought to only occur in the liver, and to 15 other amino acids, resulting in the discovery of 44 novel microbially conjugated bile acids. IMPORTANCE Our current knowledge regarding microbial bile acid transformations comes primarily from biochemical studies on a relatively small number of species or from bioinformatic predictions that rely on homology to known bile acid-transforming enzyme sequences. Therefore, much remains to be learned regarding the variety of bile acid transformations and their representation across gut microbial species. By carrying out a systematic investigation of bacterial species commonly found in the human intestinal tract, this study helps better define the gut bacteria that impact composition of the bile acid pool, which has implications in the context of metabolic disorders and cancers of the digestive tract. Our results greatly expand upon the list of bacterial species known to perform different types of bile acid transformations. This knowledge will be vital for assessing the causal connections between the microbiome, bile acid pool composition, and human health.
Keywords: bile acids; conjugation; gut bacteria; mass spectrometry; microbially conjugated bile acids; microbiome.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
