Urine tumor DNA detection of minimal residual disease in muscle-invasive bladder cancer treated with curative-intent radical cystectomy: A cohort study
- PMID: 34464379
- PMCID: PMC8407541
- DOI: 10.1371/journal.pmed.1003732
Urine tumor DNA detection of minimal residual disease in muscle-invasive bladder cancer treated with curative-intent radical cystectomy: A cohort study
Erratum in
-
Correction: Urine tumor DNA detection of minimal residual disease in muscle-invasive bladder cancer treated with curative-intent radical cystectomy: A cohort study.PLoS Med. 2021 Dec 14;18(12):e1003876. doi: 10.1371/journal.pmed.1003876. eCollection 2021 Dec. PLoS Med. 2021. PMID: 34905549 Free PMC article.
Abstract
Background: The standard of care treatment for muscle-invasive bladder cancer (MIBC) is radical cystectomy, which is typically preceded by neoadjuvant chemotherapy. However, the inability to assess minimal residual disease (MRD) noninvasively limits our ability to offer bladder-sparing treatment. Here, we sought to develop a liquid biopsy solution via urine tumor DNA (utDNA) analysis.
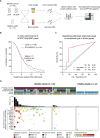
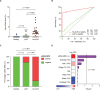
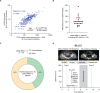
Methods and findings: We applied urine Cancer Personalized Profiling by Deep Sequencing (uCAPP-Seq), a targeted next-generation sequencing (NGS) method for detecting utDNA, to urine cell-free DNA (cfDNA) samples acquired between April 2019 and November 2020 on the day of curative-intent radical cystectomy from 42 patients with localized bladder cancer. The average age of patients was 69 years (range: 50 to 86), of whom 76% (32/42) were male, 64% (27/42) were smokers, and 76% (32/42) had a confirmed diagnosis of MIBC. Among MIBC patients, 59% (19/32) received neoadjuvant chemotherapy. utDNA variant calling was performed noninvasively without prior sequencing of tumor tissue. The overall utDNA level for each patient was represented by the non-silent mutation with the highest variant allele fraction after removing germline variants. Urine was similarly analyzed from 15 healthy adults. utDNA analysis revealed a median utDNA level of 0% in healthy adults and 2.4% in bladder cancer patients. When patients were classified as those who had residual disease detected in their surgical sample (n = 16) compared to those who achieved a pathologic complete response (pCR; n = 26), median utDNA levels were 4.3% vs. 0%, respectively (p = 0.002). Using an optimal utDNA threshold to define MRD detection, positive utDNA MRD detection was highly correlated with the absence of pCR (p < 0.001) with a sensitivity of 81% and specificity of 81%. Leave-one-out cross-validation applied to the prediction of pathologic response based on utDNA MRD detection in our cohort yielded a highly significant accuracy of 81% (p = 0.007). Moreover, utDNA MRD-positive patients exhibited significantly worse progression-free survival (PFS; HR = 7.4; 95% CI: 1.4-38.9; p = 0.02) compared to utDNA MRD-negative patients. Concordance between urine- and tumor-derived mutations, determined in 5 MIBC patients, was 85%. Tumor mutational burden (TMB) in utDNA MRD-positive patients was inferred from the number of non-silent mutations detected in urine cfDNA by applying a linear relationship derived from The Cancer Genome Atlas (TCGA) whole exome sequencing of 409 MIBC tumors. We suggest that about 58% of these patients with high inferred TMB might have been candidates for treatment with early immune checkpoint blockade. Study limitations included an analysis restricted only to single-nucleotide variants (SNVs), survival differences diminished by surgery, and a low number of DNA damage response (DRR) mutations detected after neoadjuvant chemotherapy at the MRD time point.
Conclusions: utDNA MRD detection prior to curative-intent radical cystectomy for bladder cancer correlated significantly with pathologic response, which may help select patients for bladder-sparing treatment. utDNA MRD detection also correlated significantly with PFS. Furthermore, utDNA can be used to noninvasively infer TMB, which could facilitate personalized immunotherapy for bladder cancer in the future.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: A.A.C. has patent filings related to cancer biomarkers, and has served as a consultant/advisor to Roche, Tempus, Geneoscopy, NuProbe, Daiichi Sankyo, AstraZeneca, Fenix Group International and Guidepoint; A.A.C. has stock options in Geneoscopy, research support from Roche, and ownership interests in Droplet Biosciences. V.K.A. has received research funding from ORIC Pharmaceuticals and currently serves as an employee of Bristol Myers Squibb; V.K.A. has stock options in both companies. Z.L.S. serves as a consultant/advisor for Photocure, outside of the submitted work. B.C.B. discloses honoraria from Mevion Medical Systems and consulting work for Regeneron/Sanofi, outside of the submitted work. R.K.B. is employed as a hospital medicine physician for SSM Health St. Louis. F.Q. has stock options in Centene, Gilead, and Horizon Therapeutics. No potential conflicts of interest were disclosed by the other authors.
Figures

References
-
- Sylvester RJ, van der Meijden AP, Oosterlinck W, Witjes JA, Bouffioux C, Denis L, et al.. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49(3):466–5; discussion 75–7. Epub 2006/01/31. doi: 10.1016/j.eururo.2005.12.031 . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

