Exploiting dynamic enhancer landscapes to decode macrophage and microglia phenotypes in health and disease
- PMID: 34464593
- PMCID: PMC8500948
- DOI: 10.1016/j.molcel.2021.08.004
Exploiting dynamic enhancer landscapes to decode macrophage and microglia phenotypes in health and disease
Abstract
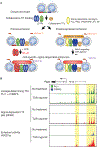
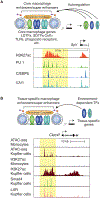
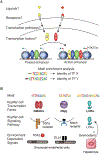
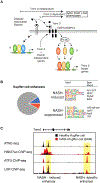
The development and functional potential of metazoan cells is dependent on combinatorial roles of transcriptional enhancers and promoters. Macrophages provide exceptionally powerful model systems for investigation of mechanisms underlying the activation of cell-specific enhancers that drive transitions in cell fate and cell state. Here, we review recent advances that have expanded appreciation of the diversity of macrophage phenotypes in health and disease, emphasizing studies of liver, adipose tissue, and brain macrophages as paradigms for other tissue macrophages and cell types. Studies of normal tissue-resident macrophages and macrophages associated with cirrhosis, obese adipose tissue, and neurodegenerative disease illustrate the major roles of tissue environment in remodeling enhancer landscapes to specify the development and functions of distinct macrophage phenotypes. We discuss the utility of quantitative analysis of environment-dependent changes in enhancer activity states as an approach to discovery of regulatory transcription factors and upstream signaling pathways.
Keywords: ATAC-seq; Alzheimer's disease; Disease associated microglia; Hematopoietic progenitor cells; Hematopoietic stem cells; Kupffer Cell; Lipid associated macrophages; Macrophage; NFkB; Nonalcoholic steatohepatitis; PU.1; Scar associated macrophages; TLR4; Trem2; enhancer; epigenetic; histone acetylation; lipopolysaccharide; microglia; obesity.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests C.K.G. is a co-founder and member of the scientific advisory board of Asteroid Therapeutics. T.D.T. and E.K. declare no competing interests.
Figures

References
-
- Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, and Tanner SD (2009). Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem 81, 6813–6822. - PubMed
-
- Bartels T, De Schepper S, and Hong S (2020). Microglia modulate neurodegeneration in Alzheimer’s and Parkinson’s diseases. Science 370, 66–69. - PubMed

