Alpha Satellite Insertion Close to an Ancestral Centromeric Region
- PMID: 34464971
- PMCID: PMC8662618
- DOI: 10.1093/molbev/msab244
Alpha Satellite Insertion Close to an Ancestral Centromeric Region
Abstract
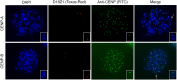
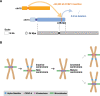
Human centromeres are mainly composed of alpha satellite DNA hierarchically organized as higher-order repeats (HORs). Alpha satellite dynamics is shown by sequence homogenization in centromeric arrays and by its transfer to other centromeric locations, for example, during the maturation of new centromeres. We identified during prenatal aneuploidy diagnosis by fluorescent in situ hybridization a de novo insertion of alpha satellite DNA from the centromere of chromosome 18 (D18Z1) into cytoband 15q26. Although bound by CENP-B, this locus did not acquire centromeric functionality as demonstrated by the lack of constriction and the absence of CENP-A binding. The insertion was associated with a 2.8-kbp deletion and likely occurred in the paternal germline. The site was enriched in long terminal repeats and located ∼10 Mbp from the location where a centromere was ancestrally seeded and became inactive in the common ancestor of humans and apes 20-25 million years ago. Long-read mapping to the T2T-CHM13 human genome assembly revealed that the insertion derives from a specific region of chromosome 18 centromeric 12-mer HOR array in which the monomer size follows a regular pattern. The rearrangement did not directly disrupt any gene or predicted regulatory element and did not alter the methylation status of the surrounding region, consistent with the absence of phenotypic consequences in the carrier. This case demonstrates a likely rare but new class of structural variation that we name "alpha satellite insertion." It also expands our knowledge on alphoid DNA dynamics and conveys the possibility that alphoid arrays can relocate near vestigial centromeric sites.
Keywords: alpha satellite; ancestral centromere; structural variation.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Alexandrov IA, Mashkova TD, Akopian TA, Medvedev LI, Kisselev LL, Mitkevich SP, Yurov YB.. 1991. Chromosome-specific alpha satellites: two distinct families on human chromosome 18. Genomics. 11(1):15–23. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. - PubMed
-
- Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M.. 1992. Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet. 89(2):247–249. - PubMed
-
- Baldini A, Ried T, Shridhar V, Ogura K, D'Aiuto L, Rocchi M, Ward DC.. 1993. An alphoid DNA sequence conserved in all human and great ape chromosomes: evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum Genet. 90(6):577–583. - PubMed

