Evolution, Mode of Transmission, and Mutational Landscape of Newly Emerging SARS-CoV-2 Variants
- PMID: 34465019
- PMCID: PMC8406297
- DOI: 10.1128/mBio.01140-21
Evolution, Mode of Transmission, and Mutational Landscape of Newly Emerging SARS-CoV-2 Variants
Abstract
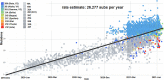
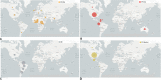
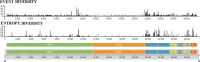
The recent emergence of multiple variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a significant concern for public health worldwide. New variants have been classified either as variants of concern (VOCs) or variants of interest (VOIs) by the CDC (USA) and WHO. The VOCs include lineages such as B.1.1.7 (20I/501Y.V1 variant), P.1 (20J/501Y.V3 variant), B.1.351 (20H/501Y.V2 variant), and B.1.617.2. In contrast, the VOI category includes B.1.525, B.1.526, P.2, and B.1.427/B.1.429. The WHO provided the alert for last two variants (P.2 and B.1.427/B.1.429) and labeled them for further monitoring. As per the WHO, these variants can be reclassified due to their status at a particular time. At the same time, the CDC (USA) has marked these two variants as VOIs up through today. This article analyzes the evolutionary patterns of all these emerging variants, as well as their geographical distributions and transmission patterns, including the circulating frequency, entropy diversity, and mutational event diversity throughout the genomes of all SARS-CoV-2 lineages. The transmission pattern was observed highest in the B.1.1.7 lineage. Our frequency evaluation found that this lineage achieved 100% frequency in early October 2020. We also critically evaluated the above emerging variants mutational landscape and significant spike protein mutations (E484K, K417T/N, N501Y, and D614G) impacting public health. Finally, the effectiveness of vaccines against newly SARS-CoV-2 variants was also analyzed. IMPORTANCE Irrespective of the aggressive vaccination drive, the newly emerging multiple SARS-CoV-2 variants are causing havoc in several countries. As per the CDC (USA) and WHO, the VOCs include the B.1.1.7, P.1, B.1.351, and B.1.617.2 lineages, while the VOIs include the B.1.525, B.1.526, P.2, and B.1.427/B.1.429 lineages. This study analyzed the evolutionary patterns, geographical distributions and transmission patterns, circulating frequency, entropy diversity, and mutational event diversity throughout the genome of significant SARS-CoV-2 lineages. A higher transmission pattern was observed for the B.1.1.7 variant. The study also evaluated the mutational landscape and important spike protein mutations (E484K, K417T/N, N501Y, and D614G) of all of the above variants. Finally, a survey was performed on the efficacy of vaccines against these variants from the previously published literature. The results presented in this article will help design future countrywide pandemic planning strategies for the emerging variants, next-generation vaccine development using alternative wild-type antigens and significant viral antigens, and immediate planning for ongoing vaccination programs worldwide.
Keywords: effect on vaccines; emerging variants; mutational landscape; transmission pattern.
Figures





















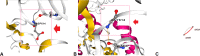
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
