Identification of a Novel FUS/ETV4 Fusion and Comparative Analysis with Other Ewing Sarcoma Fusion Proteins
- PMID: 34465585
- PMCID: PMC8568690
- DOI: 10.1158/1541-7786.MCR-21-0354
Identification of a Novel FUS/ETV4 Fusion and Comparative Analysis with Other Ewing Sarcoma Fusion Proteins
Abstract
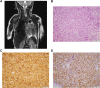
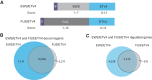
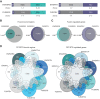
Ewing sarcoma is a pediatric bone cancer defined by a chromosomal translocation fusing one of the FET family members to an ETS transcription factor. There have been seven reported chromosomal translocations, with the most recent reported over a decade ago. We now report a novel FET/ETS translocation involving FUS and ETV4 detected in a patient with Ewing sarcoma. Here, we characterized FUS/ETV4 by performing genomic localization and transcriptional regulatory studies on numerous FET/ETS fusions in a Ewing sarcoma cellular model. Through this comparative analysis, we demonstrate significant similarities across these fusions, and in doing so, validate FUS/ETV4 as a bona fide Ewing sarcoma translocation. This study presents the first genomic comparison of Ewing sarcoma-associated translocations and reveals that the FET/ETS fusions share highly similar, but not identical, genomic localization and transcriptional regulation patterns. These data strengthen the notion that FET/ETS fusions are key drivers of, and thus pathognomonic for, Ewing sarcoma. IMPLICATIONS: Identification and initial characterization of the novel Ewing sarcoma fusion, FUS/ETV4, expands the family of Ewing fusions and extends the diagnostic possibilities for this aggressive tumor of adolescents and young adults.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures

References
-
- Riggi N, Suva ML, Stamenkovic I. Ewing's sarcoma. N Engl J Med 2021;384:154–64. - PubMed
-
- Grunewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Alava E, Kovar H, et al. Ewing sarcoma. Nat Rev Dis Primers 2018;4:5. - PubMed
-
- Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC. The ETS family of oncogenic transcription factors in solid tumours. Nat Rev Cancer 2017;17:337–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

