Acquisition of aneuploidy drives mutant p53-associated gain-of-function phenotypes
- PMID: 34465782
- PMCID: PMC8408227
- DOI: 10.1038/s41467-021-25359-z
Acquisition of aneuploidy drives mutant p53-associated gain-of-function phenotypes
Abstract
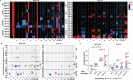
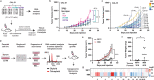
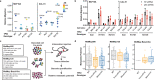
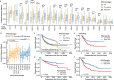
p53 is mutated in over half of human cancers. In addition to losing wild-type (WT) tumor-suppressive function, mutant p53 proteins are proposed to acquire gain-of-function (GOF) activity, leading to novel oncogenic phenotypes. To study mutant p53 GOF mechanisms and phenotypes, we genetically engineered non-transformed and tumor-derived WT p53 cell line models to express endogenous missense mutant p53 (R175H and R273H) or to be deficient for p53 protein (null). Characterization of the models, which initially differed only by TP53 genotype, revealed that aneuploidy frequently occurred in mutant p53-expressing cells. GOF phenotypes occurred clonally in vitro and in vivo, were independent of p53 alteration and correlated with increased aneuploidy. Further, analysis of outcome data revealed that individuals with aneuploid-high tumors displayed unfavorable prognoses, regardless of the TP53 genotype. Our results indicate that genetic variation resulting from aneuploidy accounts for the diversity of previously reported mutant p53 GOF phenotypes.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

