Construction of miRNA-mRNA network for the identification of key biological markers and their associated pathways in IgA nephropathy by employing the integrated bioinformatics analysis
- PMID: 34466069
- PMCID: PMC8381040
- DOI: 10.1016/j.sjbs.2021.06.079
Construction of miRNA-mRNA network for the identification of key biological markers and their associated pathways in IgA nephropathy by employing the integrated bioinformatics analysis
Abstract
Background: About half-century ago, Immunoglobulin A nephropathy (IgAN) was discovered as a complicated disease with frequent clinical symptoms. Until now, exact mechanism underlying the pathogenesis of IgAN is poorly known. Therefore, current study was aimed to understand the molecular mechanism of IgAN by identifying the key miRNAs and their targeted hub genes. The key miRNAs might contribute to the diagnosis and therapy of IgAN, and could turn out to be a new star in the field of IgAN.
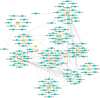
Methods: The microarray datasets were downloaded from Gene Expresssion Omnibus (GEO) database and analyzed using R package (LIMMA) in order to obtain differential expressed genes (DEGs). Then, the hub genes were identified using cytoHubba plugin of cytoscpae tool and other bioinformatics approaches including protein-protein interaction (PPI) network analysis, module analysis, and miRNA-hub gene network construction was also performed.
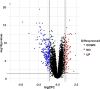
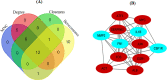
Results: A total of 348 DEGs were identified, of which 107 were upregulated genes and 241 were downregulated genes. Subsequently, the 12 overlapped genes were predicted from cytoHubba, and considered as hub genes. Moreover, a network among miRNA-hub genes was created to explore the correlation between the hub genes and their targeted miRNAs. Network construction ultimately lead to the identification of nine gene named FN1, EGR1, FOS, JUN, SERPINE1, MMP2, ATF3, MYC, and IL1B and one novel key miRNA namely, has-miR-144-3p as biomarker for diagnosis and therapy of IgAN.
Conclusion: This study updates the information and yield a new perspective in context of understanding the pathogenesis and development of IgAN. In future, key miRNAs might be capable of improving the personalized detection and therapies for IgAN. In vivo and in vitro investigation of miRNAs and pathway interaction is essential to delineate the specific roles of the novel miRNAs, which may help to further reveal the mechanisms underlying IgAN.
Keywords: BP, Cellular components; Bioinformatics analysis; CC, Molecular function; DAVID, Gene Expression Omnibus; ENCORI, Molecular Complex Detection; GEO, MicroRNA; GO, Database for annotation visualizationand integrated discovery; Gene expression profiling; Hub genes; Hub genes-miRNA network; IgAN, Differential Expressed Genes; Immunoglobulin A nephropathy; KEGG, Gene Ontology; MCODE, Search Tool for the Retrieval of Interacting Genes/Proteins; MF, Kyoto Encyclopedia of Genes and Genomes; PPI, Immunoglobulin A nephropathy; Protein-protein interaction; STRING, Biological process; miRNA, Protein-Protein Interaction.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Berger J. Intercapillary deposits of IgA-IgG. J. Urol. (Paris) 1968;74:694–695. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

