YAP regulates alveolar epithelial cell differentiation and AGER via NFIB/KLF5/NKX2-1
- PMID: 34466790
- PMCID: PMC8383002
- DOI: 10.1016/j.isci.2021.102967
YAP regulates alveolar epithelial cell differentiation and AGER via NFIB/KLF5/NKX2-1
Abstract
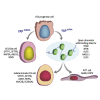
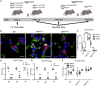
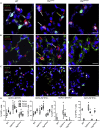
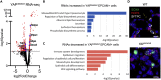
Ventilation is dependent upon pulmonary alveoli lined by two major epithelial cell types, alveolar type-1 (AT1) and 2 (AT2) cells. AT1 cells mediate gas exchange while AT2 cells synthesize and secrete pulmonary surfactants and serve as progenitor cells which repair the alveoli. We developed transgenic mice in which YAP was activated or deleted to determine its roles in alveolar epithelial cell differentiation. Postnatal YAP activation increased epithelial cell proliferation, increased AT1 cell numbers, and caused indeterminate differentiation of subsets of alveolar cells expressing atypical genes normally restricted to airway epithelial cells. YAP deletion increased expression of genes associated with mature AT2 cells. YAP activation enhanced DNA accessibility in promoters of transcription factors and motif enrichment analysis predicted target genes associated with alveolar cell differentiation. YAP participated with KLF5, NFIB, and NKX2-1 to regulate AGER. YAP plays a central role in a transcriptional network that regulates alveolar epithelial differentiation.
Keywords: Developmental genetics; Genetics; Molecular biology.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Azzolin L., Panciera T., Soligo S., Enzo E., Bicciato S., Dupont S., Bresolin S., Frasson C., Basso G., Guzzardo V. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

