Biodesulfurization Induces Reprogramming of Sulfur Metabolism in Rhodococcus qingshengii IGTS8: Proteomics and Untargeted Metabolomics
- PMID: 34468196
- PMCID: PMC8557817
- DOI: 10.1128/Spectrum.00692-21
Biodesulfurization Induces Reprogramming of Sulfur Metabolism in Rhodococcus qingshengii IGTS8: Proteomics and Untargeted Metabolomics
Abstract
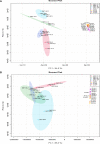
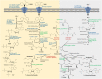
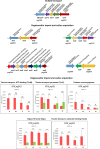
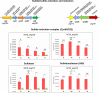
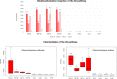
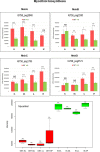
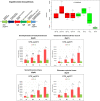
Sulfur metabolism in fuel-biodesulfurizing bacteria and the underlying physiological adaptations are not understood, which has impeded the development of a commercially viable bioprocess for fuel desulfurization. To fill these knowledge gaps, we performed comparative proteomics and untargeted metabolomics in cultures of the biodesulfurization reference strain Rhodococcus qingshengii IGTS8 grown on either inorganic sulfate or the diesel-borne organosulfur compound dibenzothiophene as a sole sulfur source. Dibenzothiophene significantly altered the biosynthesis of many sulfur metabolism proteins and metabolites in a growth phase-dependent manner, which enabled us to reconstruct the first experimental model for sulfur metabolism in a fuel-biodesulfurizing bacterium. All key pathways related to assimilatory sulfur metabolism were represented in the sulfur proteome, including uptake of the sulfur sources, sulfur acquisition, and assimilatory sulfate reduction, in addition to biosynthesis of key sulfur-containing metabolites such as S-adenosylmethionine, coenzyme A, biotin, thiamin, molybdenum cofactor, mycothiol, and ergothioneine (low-molecular weight thiols). Fifty-two proteins exhibited significantly different abundance during at least one growth phase. Sixteen proteins were uniquely detected and 47 proteins were significantly more abundant in the dibenzothiophene culture during at least one growth phase. The sulfate-free dibenzothiophene-containing culture reacted to sulfate starvation by restricting sulfur assimilation, enforcing sulfur-sparing, and maintaining redox homeostasis. Biodesulfurization triggered alternative pathways for sulfur assimilation different from those operating in the inorganic sulfate culture. Sulfur metabolism reprogramming and metabolic switches in the dibenzothiophene culture were manifested in limiting sulfite reduction and biosynthesis of cysteine, while boosting the production of methionine via the cobalamin-independent pathway, as well as the biosynthesis of the redox buffers mycothiol and ergothioneine. The omics data underscore the key role of sulfur metabolism in shaping the biodesulfurization phenotype and highlight potential targets for improving the biodesulfurization catalytic activity via metabolic engineering. IMPORTANCE For many decades, research on biodesulfurization of fossil fuels was conducted amid a large gap in knowledge of sulfur metabolism and its regulation in fuel-biodesulfurizing bacteria, which has impeded the development of a commercially viable bioprocess. In addition, lack of understanding of biodesulfurization-associated metabolic and physiological adaptations prohibited the development of efficient biodesulfurizers. Our integrated omics-based findings reveal the assimilatory sulfur metabolism in the biodesulfurization reference strain Rhodococcus qingshengii IGTS8 and show how sulfur metabolism and oxidative stress response were remodeled and orchestrated to shape the biodesulfurization phenotype. Our findings not only explain the frequently encountered low catalytic activity of native fuel-biodesulfurizing bacteria but also uncover unprecedented potential targets in sulfur metabolism that could be exploited via metabolic engineering to boost the biodesulfurization catalytic activity, a prerequisite for commercial application.
Keywords: 4S pathway; cysteine biosynthesis; dibenzothiophene; mycothiol; sulfate activation complex; sulfate starvation.
Figures

References
-
- Mohebali G, Ball AS. 2016. Biodesulfurization of diesel fuels - past, present and future perspectives. Int Biodeterior Biodegradation 110:163–180. doi:10.1016/j.ibiod.2016.03.011. - DOI
-
- Kilbane JJ. 2017. Biodesulfurization: how to make it work? Arab J Sci Eng 42:1–9. doi:10.1007/s13369-016-2269-1. - DOI
-
- Li S, Ma T. 2019. The desulfurization pathway in Rhodococcus, p 203–229. In Alvarez HM (ed), Biology of Rhodococcus; Microbiology monographs, vol 16. Springer, Berlin.
-
- Kilbane JJ. 1990. Sulfur-specific microbial metabolism of organic compounds. Resour Conserv Recycl 3:69–79. doi:10.1016/0921-3449(90)90046-7. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

