Identification of novel genetic susceptibility loci for thoracic and abdominal aortic aneurysms via genome-wide association study using the UK Biobank Cohort
- PMID: 34469433
- PMCID: PMC8409653
- DOI: 10.1371/journal.pone.0247287
Identification of novel genetic susceptibility loci for thoracic and abdominal aortic aneurysms via genome-wide association study using the UK Biobank Cohort
Abstract
Background: Thoracic aortic aneurysm (TAA) and abdominal aortic aneurysm (AAA) are known to have a strong genetic component.
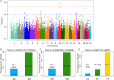
Methods and results: In a genome-wide association study (GWAS) using the UK Biobank, we analyzed the genomes of 1,363 individuals with AAA compared to 27,260 age, ancestry, and sex-matched controls (1:20 case:control study design). A similar analysis was repeated for 435 individuals with TAA compared to 8,700 controls. Polymorphism with minor allele frequency (MAF) >0.5% were evaluated. We identified novel loci near LINC01021, ATOH8 and JAK2 genes that achieved genome-wide significance for AAA (p-value <5x10-8), in addition to three known loci. For TAA, three novel loci in CTNNA3, FRMD6 and MBP achieved genome-wide significance. There was no overlap in the genes associated with AAAs and TAAs. Additionally, we identified a linkage group of high-frequency variants (MAFs ~10%) encompassing FBN1, the causal gene for Marfan syndrome, which was associated with TAA. In FinnGen PheWeb, this FBN1 haplotype was associated with aortic dissection. Finally, we found that baseline bradycardia was associated with TAA, but not AAA.
Conclusions: Our GWAS found that AAA and TAA were associated with distinct sets of genes, suggesting distinct underlying genetic architecture. We also found association between baseline bradycardia and TAA. These findings, including JAK2 association, offer plausible mechanistic and therapeutic insights. We also found a common FBN1 linkage group that is associated with TAA and aortic dissection in patients who do not have Marfan syndrome. These FBN1 variants suggest shared pathophysiology between Marfan disease and sporadic TAA.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- CDC. Aortic Aneurysm | cdc.gov. Centers for Disease Control and Prevention 2020. https://www.cdc.gov/heartdisease/aortic_aneurysm.htm. Accessed October 3, 2020.
-
- Shibamura H, Olson JM, van Vlijmen-Van Keulen C, et al.. Genome scan for familial abdominal aortic aneurysm using sex and family history as covariates suggests genetic heterogeneity and identifies linkage to chromosome 19q13. Circulation 2004;109:2103–2108. doi: 10.1161/01.CIR.0000127857.77161.A1 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

