FAN1 controls mismatch repair complex assembly via MLH1 retention to stabilize CAG repeat expansion in Huntington's disease
- PMID: 34469738
- PMCID: PMC8424649
- DOI: 10.1016/j.celrep.2021.109649
FAN1 controls mismatch repair complex assembly via MLH1 retention to stabilize CAG repeat expansion in Huntington's disease
Abstract
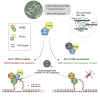
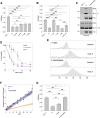
CAG repeat expansion in the HTT gene drives Huntington's disease (HD) pathogenesis and is modulated by DNA damage repair pathways. In this context, the interaction between FAN1, a DNA-structure-specific nuclease, and MLH1, member of the DNA mismatch repair pathway (MMR), is not defined. Here, we identify a highly conserved SPYF motif at the N terminus of FAN1 that binds to MLH1. Our data support a model where FAN1 has two distinct functions to stabilize CAG repeats. On one hand, it binds MLH1 to restrict its recruitment by MSH3, thus inhibiting the assembly of a functional MMR complex that would otherwise promote CAG repeat expansion. On the other hand, it promotes accurate repair via its nuclease activity. These data highlight a potential avenue for HD therapeutics in attenuating somatic expansion.
Keywords: CAG instability; DNA repair; FAN1; FAN1 nuclease activity; GWAS; Huntington’s disease; MLH1; MSH3; mismatch repair; repeat expansion.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A patent (application number 2105484.6) on the FAN1-MLH1 interaction and structural analogs for the treatment of disease has been filed by the University of Cambridge and UCL. The data presented in this patent are included in the main paper and supplemental information. G.B. is a co-founder and consultant for Adrestia Therapeutics. E.L.B. is the daughter of an advisor for Adrestia Therapeutics.
Figures



Comment in
-
SPYing on triplet repeat expansions: Insights into FAN1-MLH1 interaction and regulation.Cell Rep. 2021 Sep 14;36(11):109736. doi: 10.1016/j.celrep.2021.109736. Cell Rep. 2021. PMID: 34525375
References
-
- Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R., Nance M., Ross C.A., Scahill R.I., Wetzel R. Huntington disease. Nat. Rev. Dis. Primers. 2015;1:15005. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

