Structure of the human marker of self 5-transmembrane receptor CD47
- PMID: 34471125
- PMCID: PMC8410850
- DOI: 10.1038/s41467-021-25475-w
Structure of the human marker of self 5-transmembrane receptor CD47
Abstract
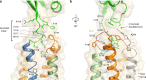
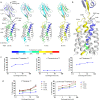
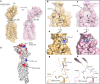
CD47 is the only 5-transmembrane (5-TM) spanning receptor of the immune system. Its extracellular domain (ECD) is a cell surface marker of self that binds SIRPα and inhibits macrophage phagocytosis, and cancer immuno-therapy approaches in clinical trials are focused on blocking CD47/SIRPα interaction. We present the crystal structure of full length CD47 bound to the function-blocking antibody B6H12. CD47 ECD is tethered to the TM domain via a six-residue peptide linker (114RVVSWF119) that forms an extended loop (SWF loop), with the fundamental role of inserting the side chains of W118 and F119 into the core of CD47 extracellular loop region (ECLR). Using hydrogen-deuterium exchange and molecular dynamics simulations we show that CD47's ECLR architecture, comprised of two extracellular loops and the SWF loop, creates a molecular environment stabilizing the ECD for presentation on the cell surface. These findings provide insights into CD47 immune recognition, signaling and therapeutic intervention.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
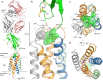



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

