A virus-free cellular model recapitulates several features of severe COVID-19
- PMID: 34471195
- PMCID: PMC8410838
- DOI: 10.1038/s41598-021-96875-7
A virus-free cellular model recapitulates several features of severe COVID-19
Abstract
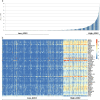
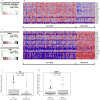
As for all newly-emergent pathogens, SARS-CoV-2 presents with a relative paucity of clinical information and experimental models, a situation hampering both the development of new effective treatments and the prediction of future outbreaks. Here, we find that a simple virus-free model, based on publicly available transcriptional data from human cell lines, is surprisingly able to recapitulate several features of the clinically relevant infections. By segregating cell lines (n = 1305) from the CCLE project on the base of their sole angiotensin-converting enzyme 2 (ACE2) mRNA content, we found that overexpressing cells present with molecular features resembling those of at-risk patients, including senescence, impairment of antibody production, epigenetic regulation, DNA repair and apoptosis, neutralization of the interferon response, proneness to an overemphasized innate immune activity, hyperinflammation by IL-1, diabetes, hypercoagulation and hypogonadism. Likewise, several pathways were found to display a differential expression between sexes, with males being in the least advantageous position, thus suggesting that the model could reproduce even the sex-related disparities observed in the clinical outcome of patients with COVID-19. Overall, besides validating a new disease model, our data suggest that, in patients with severe COVID-19, a baseline ground could be already present and, as a consequence, the viral infection might simply exacerbate a variety of latent (or inherent) pre-existing conditions, representing therefore a tipping point at which they become clinically significant.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

