ALS-related FUS mutations alter axon growth in motoneurons and affect HuD/ELAVL4 and FMRP activity
- PMID: 34471224
- PMCID: PMC8410767
- DOI: 10.1038/s42003-021-02538-8
ALS-related FUS mutations alter axon growth in motoneurons and affect HuD/ELAVL4 and FMRP activity
Abstract
Mutations in the RNA-binding protein (RBP) FUS have been genetically associated with the motoneuron disease amyotrophic lateral sclerosis (ALS). Using both human induced pluripotent stem cells and mouse models, we found that FUS-ALS causative mutations affect the activity of two relevant RBPs with important roles in neuronal RNA metabolism: HuD/ELAVL4 and FMRP. Mechanistically, mutant FUS leads to upregulation of HuD protein levels through competition with FMRP for HuD mRNA 3'UTR binding. In turn, increased HuD levels overly stabilize the transcript levels of its targets, NRN1 and GAP43. As a consequence, mutant FUS motoneurons show increased axon branching and growth upon injury, which could be rescued by dampening NRN1 levels. Since similar phenotypes have been previously described in SOD1 and TDP-43 mutant models, increased axonal growth and branching might represent broad early events in the pathogenesis of ALS.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



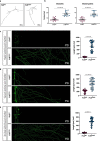
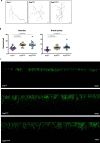
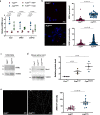
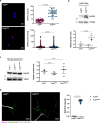
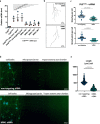
References
Publication types
MeSH terms
Substances
Grants and funding
- MR/K018523/1/MRC_/Medical Research Council/United Kingdom
- MC_EX_MR/N501931/1/MRC_/Medical Research Council/United Kingdom
- FRATTA/JAN15/946-795/MNDA_/Motor Neurone Disease Association/United Kingdom
- MR/R005184/1/MRC_/Medical Research Council/United Kingdom
- FISHER/APR14/874-792/MNDA_/Motor Neurone Disease Association/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

