A review on machine learning approaches and trends in drug discovery
- PMID: 34471498
- PMCID: PMC8387781
- DOI: 10.1016/j.csbj.2021.08.011
A review on machine learning approaches and trends in drug discovery
Abstract
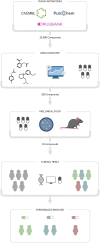
Drug discovery aims at finding new compounds with specific chemical properties for the treatment of diseases. In the last years, the approach used in this search presents an important component in computer science with the skyrocketing of machine learning techniques due to its democratization. With the objectives set by the Precision Medicine initiative and the new challenges generated, it is necessary to establish robust, standard and reproducible computational methodologies to achieve the objectives set. Currently, predictive models based on Machine Learning have gained great importance in the step prior to preclinical studies. This stage manages to drastically reduce costs and research times in the discovery of new drugs. This review article focuses on how these new methodologies are being used in recent years of research. Analyzing the state of the art in this field will give us an idea of where cheminformatics will be developed in the short term, the limitations it presents and the positive results it has achieved. This review will focus mainly on the methods used to model the molecular data, as well as the biological problems addressed and the Machine Learning algorithms used for drug discovery in recent years.
Keywords: ADMET, Absorption, distribution, metabolism, elimination and toxicity; ADR, Adverse Drug Reaction; AI, Artificial Intelligence; ANN, Artificial Neural Networks; APFP, Atom Pairs 2d FingerPrint; AUC, Area under the Curve; BBB, Blood–Brain barrier; CDK, Chemical Development Kit; CNN, Convolutional Neural Networks; CNS, Central Nervous System; CPI, Compound-protein interaction; CV, Cross Validation; Cheminformatics; DL, Deep Learning; DNA, Deoxyribonucleic acid; Deep Learning; Drug Discovery; ECFP, Extended Connectivity Fingerprints; FDA, Food and Drug Administration; FNN, Fully Connected Neural Networks; FP, Fringerprints; FS, Feature Selection; GCN, Graph Convolutional Networks; GEO, Gene Expression Omnibus; GNN, Graph Neural Networks; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MACCS, Molecular ACCess System; MCC, Matthews correlation coefficient; MD, Molecular Descriptors; MKL, Multiple Kernel Learning; ML, Machine Learning; Machine Learning; Molecular Descriptors; NB, Naive Bayes; OOB, Out of Bag; PCA, Principal Component Analyisis; QSAR; QSAR, Quantitative structure–activity relationship; RF, Random Forest; RNA, Ribonucleic Acid; SMILES, simplified molecular-input line-entry system; SVM, Support Vector Machines; TCGA, The Cancer Genome Atlas; WHO, World Health Organization; t-SNE, t-Distributed Stochastic Neighbor Embedding.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Romond E.H., Perez E.A., Bryant J., Suman V.J., Geyer C.E., Jr, Davidson N.E., Tan-Chiu E., Martino S., Paik S., Kaufman P.A. Trastuzumab plus adjuvant chemotherapy for operable her2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. - PubMed
-
- Munteanu C.R., Fernández-Blanco E., Seoane J.A., Izquierdo-Novo P., Angel Rodriguez-Fernandez J., Maria Prieto-Gonzalez J., Rabunal J.R., Pazos A. Drug discovery and design for complex diseases through qsar computational methods. Current Pharmaceutical Des. 2010;16(24):2640–2655. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
