Automated Coarse-Grained Mapping Algorithm for the Martini Force Field and Benchmarks for Membrane-Water Partitioning
- PMID: 34472843
- PMCID: PMC8444346
- DOI: 10.1021/acs.jctc.1c00322
Automated Coarse-Grained Mapping Algorithm for the Martini Force Field and Benchmarks for Membrane-Water Partitioning
Abstract
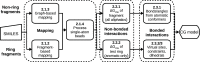
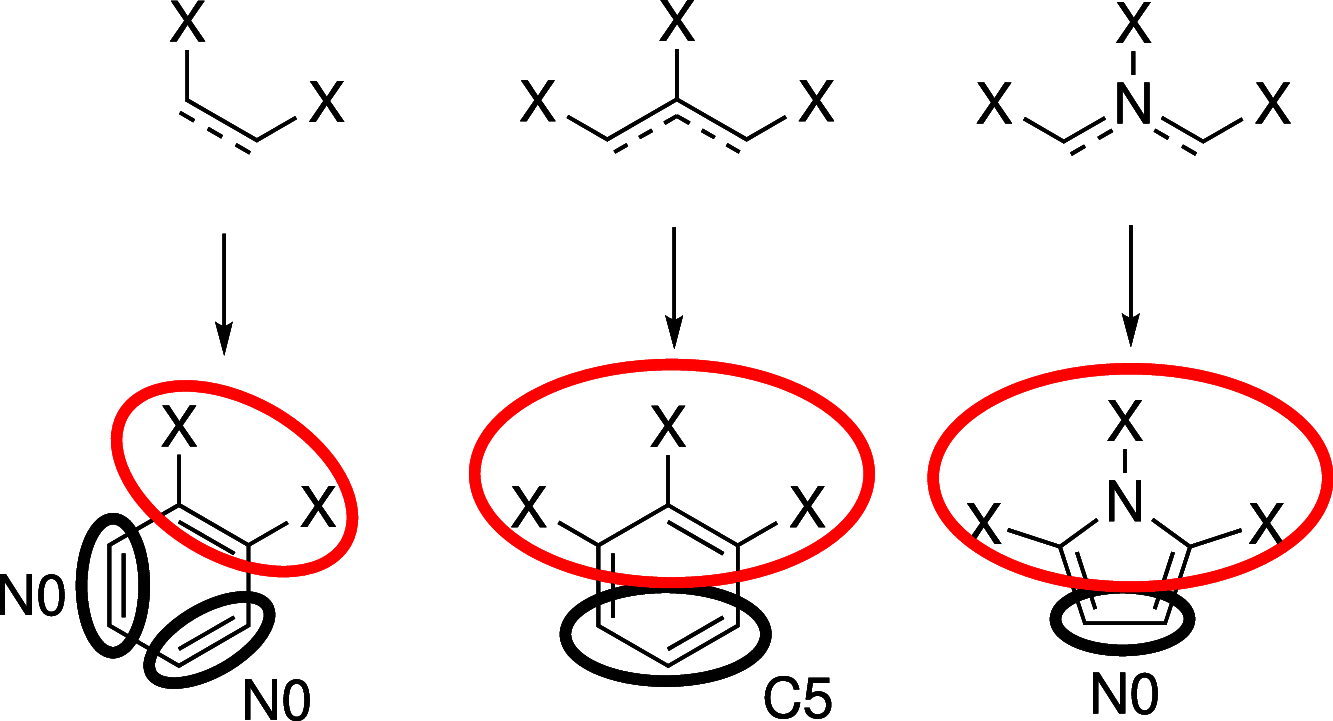
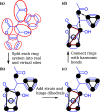
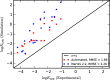
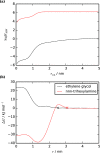
With a view to high-throughput simulations, we present an automated system for mapping and parameterizing organic molecules for use with the coarse-grained Martini force field. The method scales to larger molecules and a broader chemical space than existing schemes. The core of the mapping process is a graph-based analysis of the molecule's bonding network, which has the advantages of being fast, general, and preserving symmetry. The parameterization process pays special attention to coarse-grained beads in aromatic rings. It also includes a method for building efficient and stable frameworks of constraints for molecules with structural rigidity. The performance of the method is tested on a diverse set of 87 neutral organic molecules and the ability of the resulting models to capture octanol-water and membrane-water partition coefficients. In the latter case, we introduce an adaptive method for extracting partition coefficients from free-energy profiles to take into account the interfacial region of the membrane. We also use the models to probe the response of membrane-water partitioning to the cholesterol content of the membrane.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
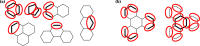







References
-
- Escher B. I.; Schwarzenbach R. P.; Westall J. C. Evaluation of Liposome–Water Partitioning of Organic Acids and Bases. 1. Development of a Sorption Model. Environ. Sci. Technol. 2000, 34, 3954–3961. 10.1021/es0010709. - DOI
-
- Seydel J. K.Drug-Membrane Interactions; John Wiley & Sons, Ltd., 2003; pp 35–50.
LinkOut - more resources
Full Text Sources

