The ATPase SRCAP is associated with the mitotic apparatus, uncovering novel molecular aspects of Floating-Harbor syndrome
- PMID: 34474679
- PMCID: PMC8414691
- DOI: 10.1186/s12915-021-01109-x
The ATPase SRCAP is associated with the mitotic apparatus, uncovering novel molecular aspects of Floating-Harbor syndrome
Abstract
Background: A variety of human genetic diseases is known to be caused by mutations in genes encoding chromatin factors and epigenetic regulators, such as DNA or histone modifying enzymes and members of ATP-dependent chromatin remodeling complexes. Floating-Harbor syndrome is a rare genetic disease affecting human development caused by dominant truncating mutations in the SRCAP gene, which encodes the ATPase SRCAP, the core catalytic subunit of the homonymous chromatin-remodeling complex. The main function of the SRCAP complex is to promote the exchange of histone H2A with the H2A.Z variant. According to the canonical role played by the SRCAP protein in epigenetic regulation, the Floating-Harbor syndrome is thought to be a consequence of chromatin perturbations. However, additional potential physiological functions of SRCAP have not been sufficiently explored.
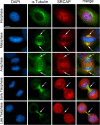
Results: We combined cell biology, reverse genetics, and biochemical approaches to study the subcellular localization of the SRCAP protein and assess its involvement in cell cycle progression in HeLa cells. Surprisingly, we found that SRCAP associates with components of the mitotic apparatus (centrosomes, spindle, midbody), interacts with a plethora of cytokinesis regulators, and positively regulates their recruitment to the midbody. Remarkably, SRCAP depletion perturbs both mitosis and cytokinesis. Similarly, DOM-A, the functional SRCAP orthologue in Drosophila melanogaster, is found at centrosomes and the midbody in Drosophila cells, and its depletion similarly affects both mitosis and cytokinesis.
Conclusions: Our findings provide first evidence suggesting that SRCAP plays previously undetected and evolutionarily conserved roles in cell division, independent of its functions in chromatin regulation. SRCAP may participate in two different steps of cell division: by ensuring proper chromosome segregation during mitosis and midbody function during cytokinesis. Moreover, our findings emphasize a surprising scenario whereby alterations in cell division produced by SRCAP mutations may contribute to the onset of Floating-Harbor syndrome.
Keywords: Cell cycle; Cytokinesis regulators; Floating-Harbor; Midbody; SRCAP.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests
Figures







References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

