Orexin receptors 1 and 2 in serotonergic neurons differentially regulate peripheral glucose metabolism in obesity
- PMID: 34475397
- PMCID: PMC8413382
- DOI: 10.1038/s41467-021-25380-2
Orexin receptors 1 and 2 in serotonergic neurons differentially regulate peripheral glucose metabolism in obesity
Abstract
The wake-active orexin system plays a central role in the dynamic regulation of glucose homeostasis. Here we show orexin receptor type 1 and 2 are predominantly expressed in dorsal raphe nucleus-dorsal and -ventral, respectively. Serotonergic neurons in ventral median raphe nucleus and raphe pallidus selectively express orexin receptor type 1. Inactivation of orexin receptor type 1 in serotonin transporter-expressing cells of mice reduced insulin sensitivity in diet-induced obesity, mainly by decreasing glucose utilization in brown adipose tissue and skeletal muscle. Selective inactivation of orexin receptor type 2 improved glucose tolerance and insulin sensitivity in obese mice, mainly through a decrease in hepatic gluconeogenesis. Optogenetic activation of orexin neurons in lateral hypothalamus or orexinergic fibers innervating raphe pallidus impaired or improved glucose tolerance, respectively. Collectively, the present study assigns orexin signaling in serotonergic neurons critical, yet differential orexin receptor type 1- and 2-dependent functions in the regulation of systemic glucose homeostasis.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

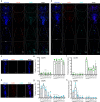




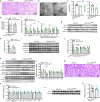
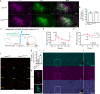
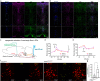
Comment in
-
Glucose control.Nat Rev Neurosci. 2021 Nov;22(11):655. doi: 10.1038/s41583-021-00530-z. Nat Rev Neurosci. 2021. PMID: 34584266 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- R21 HD098056/HD/NICHD NIH HHS/United States
- R01 DK089044/DK/NIDDK NIH HHS/United States
- R01 DK096010/DK/NIDDK NIH HHS/United States
- P50 HD105351/HD/NICHD NIH HHS/United States
- P30 DK046200/DK/NIDDK NIH HHS/United States
- P01 HL084207/HL/NHLBI NIH HHS/United States
- R01 DK108797/DK/NIDDK NIH HHS/United States
- P30 DK057521/DK/NIDDK NIH HHS/United States
- I01 BX004249/BX/BLRD VA/United States
- R01 DK075632/DK/NIDDK NIH HHS/United States
- R01 DK111401/DK/NIDDK NIH HHS/United States
- R01 NS107315/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

