Integrated Bacterial and Fungal Diversity Analysis Reveals the Gut Microbial Alterations in Diarrheic Giraffes
- PMID: 34475863
- PMCID: PMC8406688
- DOI: 10.3389/fmicb.2021.712092
Integrated Bacterial and Fungal Diversity Analysis Reveals the Gut Microbial Alterations in Diarrheic Giraffes
Abstract
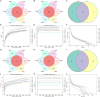
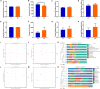
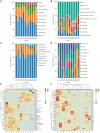
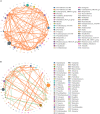
Gut microbiota has been demonstrated to be associated with multiple gastrointestinal diseases, but information regarding the gut microbial alternations in diarrheic giraffe remains scarce. Here, 16S rDNA and ITS gene amplicon sequencing were conducted to investigate the gut microbial composition and variability in diarrheic giraffes. Results demonstrated that Firmicutes and Proteobacteria were the most dominant phyla in the gut bacterial community, whereas Ascomycota and Basidiomycota were observed to be predominant in the gut fungal community regardless of health status. However, the species and relative abundance of preponderant bacterial and fungal genera in healthy and diarrheic giraffes were different. In contrast to the relatively stabilized gut fungal community, gut bacterial community displayed a significant decrease in the alpha diversity, accompanied by distinct changes in taxonomic compositions. Bacterial taxonomic analysis revealed that the relative abundances of eight phyla and 12 genera obviously increased, whereas the relative abundances of two phyla and eight genera dramatically decreased during diarrhea. Moreover, the relative richness of five fungal genera significantly increased, whereas the relative richness of seven fungal genera significantly declined in diarrheic giraffes. Taken together, this study demonstrated that diarrhea could cause significant alternations in the gut microbial composition of giraffes, and the changes in the gut bacterial community were more significant than those in the gut fungal community. Additionally, investigating the gut microbial characteristics of giraffes in different health states is beneficial to provide a theoretical basis for establishing a prevention and treatment system for diarrhea from the gut microbial perspective.
Keywords: 16S rDNA; ITS; diarrhea; giraffe; gut microbiota.
Copyright © 2021 Li, Liu, Li, He, Wang, Fakhar-e-Alam Kulyar, Li, Fu, Zhu, Wang and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

