Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria
- PMID: 34476235
- PMCID: PMC8406936
- DOI: 10.3389/fmed.2021.677720
Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria
Abstract
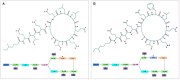
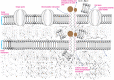
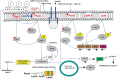
Colistin regained global interest as a consequence of the rising prevalence of multidrug-resistant Gram-negative Enterobacteriaceae. In parallel, colistin-resistant bacteria emerged in response to the unregulated use of this antibiotic. However, some Gram-negative species are intrinsically resistant to colistin activity, such as Neisseria meningitides, Burkholderia species, and Proteus mirabilis. Most identified colistin resistance usually involves modulation of lipid A that decreases or removes early charge-based interaction with colistin through up-regulation of multistep capsular polysaccharide expression. The membrane modifications occur by the addition of cationic phosphoethanolamine (pEtN) or 4-amino-l-arabinose on lipid A that results in decrease in the negative charge on the bacterial surface. Therefore, electrostatic interaction between polycationic colistin and lipopolysaccharide (LPS) is halted. It has been reported that these modifications on the bacterial surface occur due to overexpression of chromosomally mediated two-component system genes (PmrAB and PhoPQ) and mutation in lipid A biosynthesis genes that result in loss of the ability to produce lipid A and consequently LPS chain, thereafter recently identified variants of plasmid-borne genes (mcr-1 to mcr-10). It was hypothesized that mcr genes derived from intrinsically resistant environmental bacteria that carried chromosomal pmrC gene, a part of the pmrCAB operon, code three proteins viz. pEtN response regulator PmrA, sensor kinase protein PmrAB, and phosphotransferase PmrC. These plasmid-borne mcr genes become a serious concern as they assist in the dissemination of colistin resistance to other pathogenic bacteria. This review presents the progress of multiple strategies of colistin resistance mechanisms in bacteria, mainly focusing on surface changes of the outer membrane LPS structure and other resistance genetic determinants. New handier and versatile methods have been discussed for rapid detection of colistin resistance determinants and the latest approaches to revert colistin resistance that include the use of new drugs, drug combinations and inhibitors. Indeed, more investigations are required to identify the exact role of different colistin resistance determinants that will aid in developing new less toxic and potent drugs to treat bacterial infections. Therefore, colistin resistance should be considered a severe medical issue requiring multisectoral research with proper surveillance and suitable monitoring systems to report the dissemination rate of these resistant genes.
Keywords: Enterobacteriaceae; chromosomal genes; colistin resistance; electrostatic interaction; lipopolysaccharide; mcr.
Copyright © 2021 Gogry, Siddiqui, Sultan and Haq.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- WHO . Antimicrobial Resistance. Geneva: World Health Organization; (2020). Available online at: https://www.undp.org/content/undp/en/home/sustainable-development-goals.... (accessed June 20, 2021).
Publication types
LinkOut - more resources
Full Text Sources

