Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response
- PMID: 34476408
- PMCID: PMC8409526
- DOI: 10.1038/s43018-020-00110-7
Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response
Erratum in
-
Publisher Correction: Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response.Nat Cancer. 2021 Sep;2(9):994. doi: 10.1038/s43018-021-00252-2. Nat Cancer. 2021. PMID: 35121870 No abstract available.
Abstract
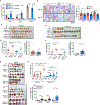
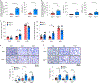
Macroautophagy (hereafter autophagy) degrades and recycles intracellular components to sustain metabolism and survival during starvation. Host autophagy promotes tumor growth by providing essential tumor nutrients. Autophagy also regulates immune cell homeostasis and function and suppresses inflammation. Although host autophagy does not promote a T-cell anti-tumor immune response in tumors with low tumor mutational burden (TMB), whether this was the case in tumors with high TMB was not known. Here we show that autophagy, especially in the liver, promotes tumor immune tolerance by enabling regulatory T-cell function and limiting stimulator of interferon genes, T-cell response and interferon-γ, which enables growth of high-TMB tumors. We have designated this as hepatic autophagy immune tolerance. Autophagy thereby promotes tumor growth through both metabolic and immune mechanisms depending on mutational load and autophagy inhibition is an effective means to promote an antitumor T-cell response in high-TMB tumors.
Conflict of interest statement
Competing interests E.W. is co-founder of Vescor Therapeutics and is a consultant for Novartis. S.G. has consulted for Roche, Merck, Novartis, Foundation Medicine and Foghorn Therapeutics. The other authors declare no competing interests. Additional information Extended data is available for this paper at https://doi.org/10.1038/s43018-020-00110-7. Supplementary information is available for this paper at https://doi.org/10.1038/s43018-020-00110-7. Reprints and permissions information is available at www.nature.com/reprints.
Figures











Comment in
-
Enhancing anti-tumor immunity by autophagy inhibition.Nat Cancer. 2021 May;2(5):484-486. doi: 10.1038/s43018-021-00214-8. Nat Cancer. 2021. PMID: 35122020 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

