Whole-genome-based phylogenomic analysis of the Belgian 2016-2017 influenza A(H3N2) outbreak season allows improved surveillance
- PMID: 34477544
- PMCID: PMC8715427
- DOI: 10.1099/mgen.0.000643
Whole-genome-based phylogenomic analysis of the Belgian 2016-2017 influenza A(H3N2) outbreak season allows improved surveillance
Abstract
Seasonal influenza epidemics are associated with high mortality and morbidity in the human population. Influenza surveillance is critical for providing information to national influenza programmes and for making vaccine composition predictions. Vaccination prevents viral infections, but rapid influenza evolution results in emerging mutants that differ antigenically from vaccine strains. Current influenza surveillance relies on Sanger sequencing of the haemagglutinin (HA) gene. Its classification according to World Health Organization (WHO) and European Centre for Disease Prevention and Control (ECDC) guidelines is based on combining certain genotypic amino acid mutations and phylogenetic analysis. Next-generation sequencing technologies enable a shift to whole-genome sequencing (WGS) for influenza surveillance, but this requires laboratory workflow adaptations and advanced bioinformatics workflows. In this study, 253 influenza A(H3N2) positive clinical specimens from the 2016-2017 Belgian season underwent WGS using the Illumina MiSeq system. HA-based classification according to WHO/ECDC guidelines did not allow classification of all samples. A new approach, considering the whole genome, was investigated based on using powerful phylogenomic tools including beast and Nextstrain, which substantially improved phylogenetic classification. Moreover, Bayesian inference via beast facilitated reassortment detection by both manual inspection and computational methods, detecting intra-subtype reassortants at an estimated rate of 15 %. Real-time analysis (i.e. as an outbreak is ongoing) via Nextstrain allowed positioning of the Belgian isolates into the globally circulating context. Finally, integration of patient data with phylogenetic groups and reassortment status allowed detection of several associations that would have been missed when solely considering HA, such as hospitalized patients being more likely to be infected with A(H3N2) reassortants, and the possibility to link several phylogenetic groups to disease severity indicators could be relevant for epidemiological monitoring. Our study demonstrates that WGS offers multiple advantages for influenza monitoring in (inter)national influenza surveillance, and proposes an improved methodology. This allows leveraging all information contained in influenza genomes, and allows for more accurate genetic characterization and reassortment detection.
Keywords: beast; influenza; next-generation sequencing; nextstrain; surveillance.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
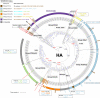
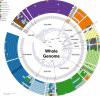

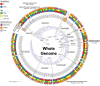
References
-
- WHO Influenza (Seasonal) Geneva: World Health Organization; 2019.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

