The protease-inhibitor SerpinB3 as a critical modulator of the stem-like subset in human cholangiocarcinoma
- PMID: 34478594
- PMCID: PMC9290104
- DOI: 10.1111/liv.15049
The protease-inhibitor SerpinB3 as a critical modulator of the stem-like subset in human cholangiocarcinoma
Abstract
Background and aims: Cholangiocarcinoma (CCA) is a form of primary liver cancer with limited therapeutic options. Recently, cancer stem cells (CSCs) have been proposed as a driving force of tumour initiation and dissemination, thus representing a crucial therapeutic target. The protease inhibitor SerpinB3 (SB3) has been identified in several malignancies including hepatocellular carcinoma. SB3 has been involved in the early events of hepatocarcinogenesis and is highly expressed in hepatic progenitor cells and in a mouse model of liver progenitor cell activation. However, only limited information on the possible role of SB3 in CCA stem-like compartment is available.
Methods: Enrichment of CCA stem-like subset was performed by sphere culture (SPH) in CCA cell lines (CCLP1, HUCCT1, MTCHC01 and SG231). Quantitative RT-PCR and Western blotting were used to detect SB3 in both SPH and parental monolayer (MON) cells. Acquired CSC-like features were analysed using an endogenous and a paracrine in vitro model, with transfection of SB3 gene or addition of recombinant SB3 to cell medium respectively. SB3 tumorigenic role was explored in an in vivo mouse model of CCA by subcutaneous injection of SB3-transfected MON (MONSB3+ ) cells in immune-deficient NOD-SCID/IL2Rgnull (NSG) mice. SB3 expression in human CCA sections was investigated by immunohistochemistry. Overall survival (OS) and time to recurrence (TTR) analyses were carried out from a transcriptome database of 104 CCA patients.
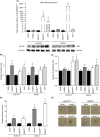
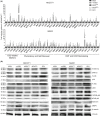
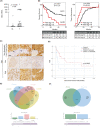
Results: SB3, barely detected in parental MON cells, was overexpressed in the same CCA cells grown as 3D SPH. Notably, MONSB3+ showed significant overexpression of genes associated with stemness (CD24, CD44, CD133), pluripotency (c-MYC, NOTCH1, STAT3, YAP, NANOG, BMI1, KLF4, OCT4, SOX2), epithelial mesenchymal transition (β-catenin, SLUG) and extracellular matrix remodelling (MMP1, MMP7, MMP9, ADAM9, ADAM10, ADAM17, ITGB3). SB3-overexpressing cells showed superior spherogenic capacity and invasion ability compared to control. Importantly, MONSB3+ exhibited activation of MAP kinases (ERK1/2, p38, JNK) as well as phosphorylation of NFκB (p65) in addition to up-regulation of the proto-oncogene β-catenin. All these effects were reversed after transient silencing of SB3. According to the in vitro finding, MONSB3+ cells retained high tumorigenic potential in NSG mice. SB3 overexpression was observed in human CCA tissues and analysis of OS as well as TTR indicated a worse prognosis in SB3+ CCA patients.
Conclusion: These findings indicate a SB3 role in mediating malignant phenotype of CCA and identify a new therapeutic target.
Keywords: SerpinB3; cancer stem cells; cholangiocarcinoma; invasion.
© 2021 The Authors. Liver International published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bertuccio P, Malvezzi M, Carioli G, et al. Reply to: "Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma". J Hepatol. 2019;71(6):1262‐1263. - PubMed
-
- Banales JM, Cardinale V, Carpino G, et al. Fouassier L and others. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS‐CCA). Nat Rev Gastroenterol Hepatol. 2016;13(5):261‐280. - PubMed
-
- Oikawa T. Cancer Stem cells and their cellular origins in primary liver and biliary tract cancers. Hepatology. 2016;64(2):645‐651. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

