Engineered 3D vessel-on-chip using hiPSC-derived endothelial- and vascular smooth muscle cells
- PMID: 34478648
- PMCID: PMC8452600
- DOI: 10.1016/j.stemcr.2021.08.003
Engineered 3D vessel-on-chip using hiPSC-derived endothelial- and vascular smooth muscle cells
Abstract
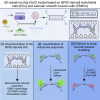
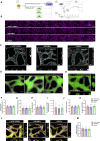
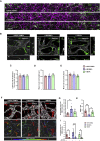
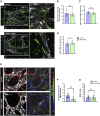
Crosstalk between endothelial cells (ECs) and pericytes or vascular smooth muscle cells (VSMCs) is essential for the proper functioning of blood vessels. This balance is disrupted in several vascular diseases but there are few experimental models which recapitulate this vascular cell dialogue in humans. Here, we developed a robust multi-cell type 3D vessel-on-chip (VoC) model based entirely on human induced pluripotent stem cells (hiPSCs). Within a fibrin hydrogel microenvironment, the hiPSC-derived vascular cells self-organized to form stable microvascular networks reproducibly, in which the vessels were lumenized and functional, responding as expected to vasoactive stimulation. Vascular organization and intracellular Ca2+ release kinetics in VSMCs could be quantified using automated image analysis based on open-source software CellProfiler and ImageJ on widefield or confocal images, setting the stage for use of the platform to study vascular (patho)physiology and therapy.
Keywords: 3D vessel-on-chip; VoC; functional readouts; hiPSC-ECs; hiPSC-VSMCs; hiPSC-derived endothelial cells; hiPSC-derived vascular smooth muscle cells; microfluidics; organ-on-chip; vessels-on-chip.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Cochrane A., Albers H.J., Passier R., Mummery C.L., van den Berg A., Orlova V.V., van der Meer A.D. Advanced in vitro models of vascular biology: human induced pluripotent stem cells and organ-on-chip technology. Adv. Drug Deliv. Rev. 2019;140:68–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

