TXNIP interaction with GLUT1 depends on PI(4,5)P2
- PMID: 34478732
- PMCID: PMC8464517
- DOI: 10.1016/j.bbamem.2021.183757
TXNIP interaction with GLUT1 depends on PI(4,5)P2
Abstract
GLUT1 is a major glucose facilitator expressed ubiquitously among tissues. Upregulation of its expression plays an important role in the development of many types of cancer and metabolic diseases. Thioredoxin-interacting protein (TXNIP) is an α-arrestin that acts as an adaptor for GLUT1 in clathrin-mediated endocytosis. It regulates cellular glucose uptake in response to both intracellular and extracellular signals via its control on GLUT1-4. In order to understand the interaction between GLUT1 and TXNIP, we generated GLUT1 lipid nanodiscs and carried out isothermal titration calorimetry and single-particle electron microscopy experiments. We found that GLUT1 lipid nanodiscs and TXNIP interact in a 1:1 ratio and that this interaction requires phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2 or PIP2).
Keywords: Electron microscopy; GLUT1; Glucose metabolism; Nanodiscs; PI(4,5)P(2); TXNIP (thioredoxin-interacting protein).
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
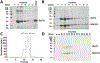




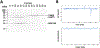
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

