An overview of genes and mutations associated with Chlamydiae species' resistance to antibiotics
- PMID: 34479551
- PMCID: PMC8414684
- DOI: 10.1186/s12941-021-00465-4
An overview of genes and mutations associated with Chlamydiae species' resistance to antibiotics
Abstract
Background: Chlamydiae are intracellular bacteria that cause various severe diseases in humans and animals. The common treatment for chlamydia infections are antibiotics. However, when antibiotics are misused (overuse or self-medication), this may lead to resistance of a number of chlamydia species, causing a real public health problem worldwide.
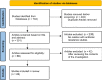
Materials and methods: In the present work, a comprehensive literature search was conducted in the following databases: PubMed, Google Scholar, Cochrane Library, Science direct and Web of Science. The primary purpose is to analyse a set of data describing the genes and mutations involved in Chlamydiae resistance to antibiotic mechanisms. In addition, we proceeded to a filtration process among 704 retrieved articles, then finished by focusing on 24 studies to extract data that met our requirements.
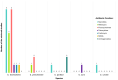
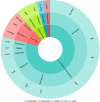
Results: The present study revealed that Chlamydia trachomatis may develop resistance to macrolides via mutations in the 23S rRNA, rplD, rplV genes, to rifamycins via mutations in the rpoB gene, to fluoroquinolones via mutations in the gyrA, parC and ygeD genes, to tetracyclines via mutations in the rpoB gene, to fosfomycin via mutations in the murA gene, to MDQA via mutations in the secY gene. Whereas, Chlamydia pneumoniae may develop resistance to rifamycins via mutations in the rpoB gene, to fluoroquinolones via mutations in the gyrA gene. Furthermore, the extracted data revealed that Chlamydia psittaci may develop resistance to aminoglycosides via mutations in the 16S rRNA and rpoB genes, to macrolides via mutations in the 23S rRNA gene. Moreover, Chlamydia suis can become resistance to tetracyclines via mutations in the tet(C) gene. In addition, Chlamydia caviae may develop resistance to macrolides via variations in the 23S rRNA gene. The associated mechanisms of resistance are generally: the inhibition of bacteria's protein synthesis, the inhibition of bacterial enzymes' action and the inhibition of bacterial transcription process.
Conclusion: This literature review revealed the existence of diverse mutations associated with resistance to antibiotics using molecular tools and targeting chlamydia species' genes. Furthermore, these mutations were shown to be associated with different mechanisms that led to resistance. In that regards, more mutations and information can be shown by a deep investigation using the whole genome sequencing. Certainly, this can help improving to handle chlamydia infections and healthcare improvement by decreasing diseases complications and medical costs.
Keywords: Antibiotics; Chlamydiae; Dataset; Genes; Infections; Mutations; Resistance.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Mutations in a 23S rRNA gene of Chlamydia trachomatis associated with resistance to macrolides.Antimicrob Agents Chemother. 2004 Apr;48(4):1347-9. doi: 10.1128/AAC.48.4.1347-1349.2004. Antimicrob Agents Chemother. 2004. PMID: 15047540 Free PMC article.
-
Touchdown enzyme time release-PCR for detection and identification of Chlamydia trachomatis, C. pneumoniae, and C. psittaci using the 16S and 16S-23S spacer rRNA genes.J Clin Microbiol. 2000 Mar;38(3):1085-93. doi: 10.1128/JCM.38.3.1085-1093.2000. J Clin Microbiol. 2000. PMID: 10699002 Free PMC article.
-
Macrolide and fluoroquinolone resistance is uncommon in clinical strains of Chlamydia trachomatis.J Infect Chemother. 2018 Aug;24(8):610-614. doi: 10.1016/j.jiac.2018.03.007. Epub 2018 Apr 4. J Infect Chemother. 2018. PMID: 29627327
-
Molecular mechanisms of Chlamydia trachomatis resistance to antimicrobial drugs.Front Biosci (Landmark Ed). 2018 Jan 1;23(4):656-670. doi: 10.2741/4611. Front Biosci (Landmark Ed). 2018. PMID: 28930567 Review.
-
Mycoplasma pneumoniae: susceptibility and resistance to antibiotics.Future Microbiol. 2011 Apr;6(4):423-31. doi: 10.2217/fmb.11.18. Future Microbiol. 2011. PMID: 21526943 Review.
Cited by
-
Adoption of an in-silico analysis approach to assess the functional and structural impacts of rpoB-encoded protein mutations on Chlamydia pneumoniae sensitivity to antibiotics.BMC Microbiol. 2025 Mar 19;25(1):157. doi: 10.1186/s12866-025-03860-5. BMC Microbiol. 2025. PMID: 40102727 Free PMC article.
-
Alternative strategies for Chlamydia treatment: Promising non-antibiotic approaches.Front Microbiol. 2022 Nov 23;13:987662. doi: 10.3389/fmicb.2022.987662. eCollection 2022. Front Microbiol. 2022. PMID: 36504792 Free PMC article. Review.
-
An Atypical Pneumonia Case of Quinolone-Refractory Chlamydia Pneumoniae Successfully Treated With Omadacycline.Infect Drug Resist. 2025 May 7;18:2357-2363. doi: 10.2147/IDR.S522259. eCollection 2025. Infect Drug Resist. 2025. PMID: 40357417 Free PMC article.
-
An in silico analysis of rpoB mutations to affect Chlamydia trachomatis sensitivity to rifamycin.J Genet Eng Biotechnol. 2022 Oct 25;20(1):146. doi: 10.1186/s43141-022-00428-y. J Genet Eng Biotechnol. 2022. PMID: 36282371 Free PMC article.
-
Chlamydial and Gonococcal Genital Infections: A Narrative Review.J Pers Med. 2023 Jul 21;13(7):1170. doi: 10.3390/jpm13071170. J Pers Med. 2023. PMID: 37511783 Free PMC article. Review.
References
-
- Radouani F, Takourt B, Benomar H, Guerbaoui M, Bekkay M, Boutaleb Y, et al. Chlamydia infection and female low fertility in Morocco. Pathol Biol. 1997;45(6):491–5. - PubMed
-
- Takourt B, de Barbeyrac B, Khyatti M, Radouani F, Bebear C, Dessus-Babus S, et al. Direct genotyping and nucleotide sequence analysis of VS1 and VS2 of the Omp1 gene of Chlamydia trachomatis from Moroccan trachomatous specimens. Microbes Infect. 2001;3(6):459–466. doi: 10.1016/s1286-4579(01)01401-0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous