Mass cytometry profiling of human dendritic cells in blood and tissues
- PMID: 34480131
- PMCID: PMC10538357
- DOI: 10.1038/s41596-021-00599-x
Mass cytometry profiling of human dendritic cells in blood and tissues
Abstract
The immune system comprises distinct functionally specialized cell populations, which can be characterized in depth by mass cytometry protein profiling. Unfortunately, the low-throughput nature of mass cytometry has made it challenging to analyze minor cell populations. This is the case for dendritic cells, which represent 0.2-2% of all immune cells in tissues and yet perform the critical task of initiating and modulating immune responses. Here, we provide an optimized step-by-step protocol for the characterization of well-known and emerging human dendritic cell populations in blood and tissues using mass cytometry. We provide detailed instructions for the generation of single-cell suspensions, sample enrichment, staining, acquisition and data analysis. We also include a barcoding option that reduces acquisition variability and allows the analysis of low numbers of dendritic cells, i.e., ~20,000. In contrast to other protocols, we emphasize the use of negative selection approaches to enrich for minor populations of immune cells while avoiding their activation. The entire procedure can be completed in 2-3 d and can be conveniently paused at several stages. The procedure described in this robust and reliable protocol allows the analysis of human dendritic cells in health and disease and during vaccination.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures


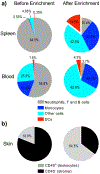


References
-
- Steinman RM & Idoyaga J Features of the dendritic cell lineage. Immunol Rev 234, 5–17 (2010). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

