Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis
- PMID: 34480146
- PMCID: PMC8484604
- DOI: 10.1038/s41423-021-00760-2
Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis
Abstract
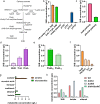
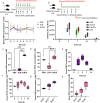
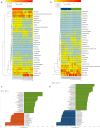
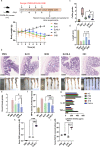
Colitis is a common disease of the colon that is very difficult to treat. Probiotic bacteria could be an effective treatment. The probiotic Escherichia coli Nissle 1917 (EcN) was engineered to synthesize the ketone body (R)-3-hydroxybutyrate (3HB) for sustainable production in the gut lumen of mice suffering from colitis. Components of heterologous 3HB synthesis routes were constructed, expressed, optimized, and inserted into the EcN genome, combined with deletions in competitive branch pathways. The genome-engineered EcN produced the highest 3HB level of 0.6 g/L under microaerobic conditions. The live therapeutic was found to colonize the mouse gastrointestinal tract over 14 days, elevating gut 3HB and short-chain-length fatty acid (SCFA) levels 8.7- and 3.1-fold compared to those of wild-type EcN, respectively. The sustainable presence of 3HB in mouse guts promoted the growth of probiotic bacteria, especially Akkermansia spp., to over 31% from the initial 2% of all the microbiome. As a result, the engineered EcN termed EcNL4 ameliorated colitis induced via dextran sulfate sodium (DSS) in mice. Compared to wild-type EcN or oral administration of 3HB, oral EcNL4 uptake demonstrated better effects on mouse weights, colon lengths, occult blood levels, gut tissue myeloperoxidase activity and proinflammatory cytokine concentrations. Thus, a promising live bacterium was developed to improve colonic microenvironments and further treat colitis. This proof-of-concept design can be employed to treat other diseases of the colon.
Keywords: 3HB; Colitis; Colonic microenvironment; Escherichia coli Nissle 1917; Metabolic engineering; Microbiomes; Probiotics; Synthetic biology; β-hydroxybutyrate.
© 2021. The Author(s), under exclusive licence to CSI and USTC.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- 2020Z99CFG002/Tsinghua University (THU)
- 21761132013/National Natural Science Foundation of China (National Science Foundation of China)
- 31771886/National Natural Science Foundation of China (National Science Foundation of China)
- 31971170/National Natural Science Foundation of China (National Science Foundation of China)
- 31870859/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

