The genomics of heart failure: design and rationale of the HERMES consortium
- PMID: 34480422
- PMCID: PMC8712846
- DOI: 10.1002/ehf2.13517
The genomics of heart failure: design and rationale of the HERMES consortium
Abstract
Aims: The HERMES (HEart failure Molecular Epidemiology for Therapeutic targetS) consortium aims to identify the genomic and molecular basis of heart failure.
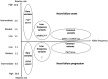
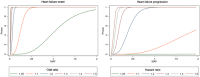
Methods and results: The consortium currently includes 51 studies from 11 countries, including 68 157 heart failure cases and 949 888 controls, with data on heart failure events and prognosis. All studies collected biological samples and performed genome-wide genotyping of common genetic variants. The enrolment of subjects into participating studies ranged from 1948 to the present day, and the median follow-up following heart failure diagnosis ranged from 2 to 116 months. Forty-nine of 51 individual studies enrolled participants of both sexes; in these studies, participants with heart failure were predominantly male (34-90%). The mean age at diagnosis or ascertainment across all studies ranged from 54 to 84 years. Based on the aggregate sample, we estimated 80% power to genetic variant associations with risk of heart failure with an odds ratio of ≥1.10 for common variants (allele frequency ≥ 0.05) and ≥1.20 for low-frequency variants (allele frequency 0.01-0.05) at P < 5 × 10-8 under an additive genetic model.
Conclusions: HERMES is a global collaboration aiming to (i) identify the genetic determinants of heart failure; (ii) generate insights into the causal pathways leading to heart failure and enable genetic approaches to target prioritization; and (iii) develop genomic tools for disease stratification and risk prediction.
Keywords: Association studies; Biomarkers; Cardiomyopathy; Genetics; Heart failure.
© 2021 The Authors. ESC Heart Failure published by John Wiley & Sons Ltd on behalf of European Society of Cardiology.
Conflict of interest statement
Daniel I. Swerdlow is an employee of Silence Therapeutics plc. Joshua D. Backman and Jonathan H. Chung are employees of Regeneron Genetics Center. Simon de Denus was supported through grants from Pfizer, AstraZeneca, Roche Molecular Science, DalCor, and Novartis. Bruce M. Psaty serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. Carolina Roselli is supported by a grant from Bayer AG to the Broad Institute focused on the development of therapeutics for cardiovascular disease. Jean‐Claude Tardif has received research support from Amarin, AstraZeneca, DalCor, Ionis, Pfizer, RegenexBio, Sanofi, and Servier and honoraria from AstraZeneca, DalCor, Pfizer, Sanofi, and Servier; holds minor equity interest in DalCor; and is an author of a patent on pharmacogenomics‐guided CETP inhibition. Benoit Tyl receives full‐time salary from Servier. Harvey D. White reports grants and personal fees from Eli Lilly and Company, Omthera Pharmaceuticals, Pfizer USA, Eisai Inc., DalCor Pharma UK Inc, CSL Behring LLC, American Regent, Sanofi‐Aventis Australia Pty Ltd, and Esperion Therapeutics Inc. and personal fees from Genentech, Inc., outside the submitted work. Steven A. Lubitz receives sponsored research support from Bristol Myers Squibb/Pfizer, Bayer AG, Boehringer Ingelheim, and Fitbit and has consulted for Bristol Myers Squibb/Pfizer and Bayer AG. Michael E. Dunn is an employee of Regeneron Pharmaceuticals. Marie‐Pierre Dubé has received honoraria from Dalcor, holds minor equity interest in DalCor, is an author of a patent on pharmacogenomics‐guided CETP inhibition, and has received research support (access to samples and data) from AstraZeneca, Pfizer, Servier, Sanofi, and GlaxoSmithKline. Authors affiliated with deCODE genetics are employed by deCODE genetics/Amgen Inc.
Figures


References
-
- Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JG, Coats AJ, Falk V, González‐Juanatey JR, Harjola VP, Jankowska EA, Jessup M, Linde C, Nihoyannopoulos P, Parissis JT, Pieske B, Riley JP, Rosano GM, Ruilope LM, Ruschitzka F, Rutten FH, van der Meer P, Authors/Task Force Members . Document Reviewers 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC)Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail 2016; 18: 891–975. - PubMed
Publication types
MeSH terms
Grants and funding
- U10 HL110309/HL/NHLBI NIH HHS/United States
- R01 HL139731/HL/NHLBI NIH HHS/United States
- U10 HL110262/HL/NHLBI NIH HHS/United States
- 75N92019D00031/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- U10 HL110336/HL/NHLBI NIH HHS/United States
- K08 HL153950/HL/NHLBI NIH HHS/United States
- MC_UU_00006/1/MRC_/Medical Research Council/United Kingdom
- MR/S003754/1/MRC_/Medical Research Council/United Kingdom
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- U01 DK062413/DK/NIDDK NIH HHS/United States
- DH_/Department of Health/United Kingdom
- U10 HL110342/HL/NHLBI NIH HHS/United States
- RE/18/6/34217/BHF_/British Heart Foundation/United Kingdom
- U10 HL084904/HL/NHLBI NIH HHS/United States
- U10 HL110338/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- U10 HL110302/HL/NHLBI NIH HHS/United States
- MR/K006584/1/MRC_/Medical Research Council/United Kingdom
- U10 HL110337/HL/NHLBI NIH HHS/United States
- MC_PC_13041/MRC_/Medical Research Council/United Kingdom
- G0902393/MRC_/Medical Research Council/United Kingdom
- UL1 TR001881/TR/NCATS NIH HHS/United States
- U10 HL110312/HL/NHLBI NIH HHS/United States
- R01 HL141232/HL/NHLBI NIH HHS/United States
- U10 HL110297/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

