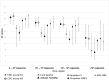
Comparison of diagnostic performance of five molecular assays for detection of SARS-CoV-2
- PMID: 34481324
- PMCID: PMC8343369
- DOI: 10.1016/j.diagmicrobio.2021.115518
Comparison of diagnostic performance of five molecular assays for detection of SARS-CoV-2
Abstract
We compared the performance of the Abbott Real Time SARS-CoV-2 assay (Abbott assay), Aptima™ SARS-CoV-2 assay (Aptima assay), BGI Real-Time SARS-CoV-2 assay (BGI assay), Lyra® SARS-CoV-2 assay (Lyra assay), and DiaSorin Simplexa™ COVID assay for SARS-CoV-2 detection. Residual nasopharyngeal samples (n = 201) submitted for routine SARS-CoV-2 testing by Simplexa assay during June-July 2020 and January 2021 were salvaged. Aliquots were tested on other assays and compared against the CDC 2019-nCoV Real-Time RT-PCR assay. Viral load in positive samples was determined by droplet digital PCR. Among 201 samples, 99 were positive and 102 were negative by the CDC assay. The Aptima and Abbott assays exhibited the highest positive percent agreement (PPA) at 98.9% while the BGI assay demonstrated the lowest PPA of 89.9% with 10 missed detections. Negative percent agreement for all 5 platforms was comparable, ranging from 96.1% to 100%. The performance of all five assays was comparable.
Keywords: COVID-19; Molecular assay comparison; SARS-CoV-2 detection.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors report no conflicts of interest relevant to this article.
Figures
References
-
- Hanson KE, Arias CA, Englund JA, Lee MJ, Loeb M, Patel R, et al. IDSA; 2020. Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19.https://www.idsociety.org/practice-guideline/covid-19-guideline-diagnost... AMC. Available at: Accessed 07/15/2020. - PMC - PubMed
-
- AMP . Association for Molecular Pathology; 2020. SARS-CoV-2 molecular testing: summary of recent SARS-CoV-2 molecular testing survey.https://www.amp.org/advocacy/sars-cov-2-survey/ Available at: Accessed 07/15/2020.
-
- Araj GF BM, Bizri NA, Ghizzawi L. Testing for COVID-19: when, who, and what test? Lebanese Med J. 2020;68:16–26.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous