A novel air-dried multiplex high-resolution melt assay for the detection of extended-spectrum β-lactamase and carbapenemase genes
- PMID: 34482019
- PMCID: PMC8692233
- DOI: 10.1016/j.jgar.2021.08.006
A novel air-dried multiplex high-resolution melt assay for the detection of extended-spectrum β-lactamase and carbapenemase genes
Abstract
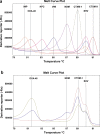
Objectives: This study aimed to develop and evaluate a novel air-dried high-resolution melt (HRM) assay to detect eight major extended-spectrum β-lactamase (ESBL) (blaSHV and blaCTX-M groups 1 and 9) and carbapenemase (blaNDM, blaIMP, blaKPC, blaVIM and blaOXA-48-like) genes that confer resistance to cephalosporins and carbapenems.
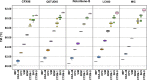
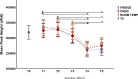
Methods: The assay was evaluated using 439 DNA samples extracted from bacterial isolates from Nepal, Malawi and the UK and 390 clinical isolates from Nepal with known antimicrobial susceptibility. Assay reproducibility was evaluated across five different real-time quantitative PCR (qPCR) instruments [Rotor-Gene® Q, QuantStudioTM 5, CFX96, LightCycler® 480 and Magnetic Induction Cycler (Mic)]. Assay stability was also assessed under different storage temperatures (6.2 ± 0.9°C, 20.4 ± 0.7°C and 29.7 ± 1.4°C) at six time points over 8 months.
Results: The sensitivity and specificity (with 95% confidence intervals) for detecting ESBL and carbapenemase genes was 94.7% (92.5-96.5%) and 99.2% (98.8-99.5%) compared with the reference gel-based PCR and sequencing and 98.3% (97.0-99.3%) and 98.5% (98.0-98.9%) compared with the original HRM wet PCR mix format. Overall agreement was 91.1% (90.0-92.9%) when predicting phenotypic resistance to cefotaxime and meropenem among Enterobacteriaceae isolates. We observed almost perfect inter-machine reproducibility of the air-dried HRM assay, and no loss of sensitivity occurred under all storage conditions and time points.
Conclusion: We present a ready-to-use air-dried HRM PCR assay that offers an easy, thermostable, fast and accurate tool for the detection of ESBL and carbapenemase genes in DNA samples to improve antimicrobial resistance detection.
Keywords: Antimicrobial resistance; Carbapenemase; ESBL; Extended-spectrum β-lactamase; High-resolution melting; Molecular diagnostics.
Copyright © 2021 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures

Similar articles
-
Detection of carbapenemases blaOXA48-blaKPC-blaNDM-blaVIM and extended-spectrum-β-lactamase blaOXA1-blaSHV-blaTEM genes in Gram-negative bacterial isolates from ICU burns patients.Ann Clin Microbiol Antimicrob. 2022 May 19;21(1):18. doi: 10.1186/s12941-022-00510-w. Ann Clin Microbiol Antimicrob. 2022. PMID: 35590320 Free PMC article.
-
Multiplex real-time PCR assay for the detection of extended-spectrum β-lactamase and carbapenemase genes using melting curve analysis.J Microbiol Methods. 2016 May;124:72-8. doi: 10.1016/j.mimet.2016.03.014. Epub 2016 Mar 25. J Microbiol Methods. 2016. PMID: 27021662
-
Carbapenemase and ESBL genes with class 1 integron among fermenting and nonfermenting bacteria isolated from water sources from India.Lett Appl Microbiol. 2020 Jul;71(1):70-77. doi: 10.1111/lam.13228. Epub 2019 Nov 8. Lett Appl Microbiol. 2020. PMID: 31587338
-
High prevalence of carbapenemase, AmpC β-lactamase and aminoglycoside resistance genes in extended-spectrum β-lactamase-positive uropathogens from Northern India.J Glob Antimicrob Resist. 2020 Mar;20:197-203. doi: 10.1016/j.jgar.2019.07.029. Epub 2019 Aug 6. J Glob Antimicrob Resist. 2020. PMID: 31398493
-
A highly multiplexed melt-curve assay for detecting the most prevalent carbapenemase, ESBL, and AmpC genes.Diagn Microbiol Infect Dis. 2020 Aug;97(4):115076. doi: 10.1016/j.diagmicrobio.2020.115076. Epub 2020 May 8. Diagn Microbiol Infect Dis. 2020. PMID: 32521424
Cited by
-
Decoding human cytomegalovirus for the development of innovative diagnostics to detect congenital infection.Pediatr Res. 2024 Jan;95(2):532-542. doi: 10.1038/s41390-023-02957-9. Epub 2023 Dec 25. Pediatr Res. 2024. PMID: 38146009 Free PMC article. Review.
-
Molecular surveillance reveals widespread colonisation by carbapenemase and extended spectrum beta-lactamase producing organisms in neonatal units in Kenya and Nigeria.Antimicrob Resist Infect Control. 2023 Feb 22;12(1):14. doi: 10.1186/s13756-023-01216-0. Antimicrob Resist Infect Control. 2023. PMID: 36814315 Free PMC article.
-
The correlation between antibiotic usage and antibiotic resistance: a 3-year retrospective study.Front Cell Infect Microbiol. 2025 Aug 1;15:1608921. doi: 10.3389/fcimb.2025.1608921. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40822587 Free PMC article.
References
-
- World Health Organization (WHO) WHO; Geneva, Switzerland: 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics.
-
- O'Neill J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Review on Antimicrobial Resistance; 2016
-
- Kerremans JJ, Verboom P, Stijnen T, Hakkaart-van Roijen L, Goessens W, Verbrugh HA, et al. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J Antimicrob Chemother. 2008;61:428–435. doi: 10.1093/jac/dkm497. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

