Phylotaxogenomics for the Reappraisal of the Genus Roseomonas With the Creation of Six New Genera
- PMID: 34484138
- PMCID: PMC8414978
- DOI: 10.3389/fmicb.2021.677842
Phylotaxogenomics for the Reappraisal of the Genus Roseomonas With the Creation of Six New Genera
Abstract
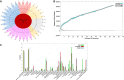
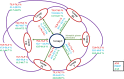
The genus Roseomonas is a significant group of bacteria which is invariably of great clinical and ecological importance. Previous studies have shown that the genus Roseomonas is polyphyletic in nature. Our present study focused on generating a lucid understanding of the phylogenetic framework for the re-evaluation and reclassification of the genus Roseomonas. Phylogenetic studies based on the 16S rRNA gene and 92 concatenated genes suggested that the genus is heterogeneous, forming seven major groups. Existing Roseomonas species were subjected to an array of genomic, phenotypic, and chemotaxonomic analyses in order to resolve the heterogeneity. Genomic similarity indices (dDDH and ANI) indicated that the members were well-defined at the species level. The Percentage of Conserved Proteins (POCP) and the average Amino Acid Identity (AAI) values between the groups of the genus Roseomonas and other interspersing members of the family Acetobacteraceae were below 65 and 70%, respectively. The pan-genome evaluation depicted that the pan-genome was an open type and the members shared 958 core genes. This claim of reclassification was equally supported by the phenotypic and chemotaxonomic differences between the groups. Thus, in this study, we propose to re-evaluate and reclassify the genus Roseomonas and propose six novel genera as Pararoseomonas gen. nov., Falsiroseomonas gen. nov., Paeniroseomonas gen. nov., Plastoroseomonas gen. nov., Neoroseomonas gen. nov., and Pseudoroseomonas gen. nov.
Keywords: Roseomonas; average amino acid Identity (AAI); percentage of conserved proteins (POCP); phylotaxogenomics; reclassification.
Copyright © 2021 Rai, Jagadeeshwari, Deepshikha, Smita, Sasikala and Ramana.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Andreeva I. S., Pechurkina N. I., Morozova O. V., Riabchikova E. I., Belikov S. I., Puchkova L. I., et al. (2007). The new eubacterium Roseomonas baikalica sp. nov. isolated from core samples collected by deep-hole drilling of the bottom of Lake Baikal. Mikrobiologiia 76 552–559. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

